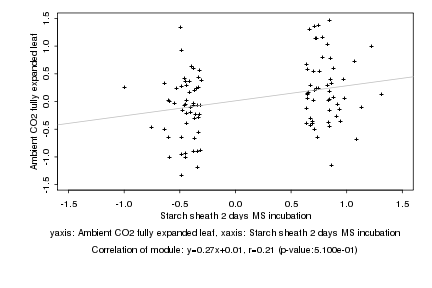
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Starch sheath 2 days MS incubation |
Log2 signal ratio
Ambient CO2 fully expanded leaf |
| 1 |
254832_at |
AT4G12490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.304 |
0.141 |
| 2 |
256787_at |
AT3G13790
|
ATBFRUCT1, ATCWINV1, ARABIDOPSIS THALIANA CELL WALL INVERTASE 1 |
1.221 |
0.996 |
| 3 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
1.126 |
-0.094 |
| 4 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
1.086 |
-0.676 |
| 5 |
256825_at |
AT3G22120
|
CWLP, cell wall-plasma membrane linker protein |
1.065 |
0.734 |
| 6 |
251899_at |
AT3G54400
|
[Eukaryotic aspartyl protease family protein] |
0.978 |
0.062 |
| 7 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
0.964 |
0.399 |
| 8 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
0.936 |
-0.345 |
| 9 |
256890_at |
AT3G23830
|
AtGRP4, GR-RBP4, GRP4, glycine-rich RNA-binding protein 4 |
0.931 |
-0.135 |
| 10 |
263380_at |
AT2G40200
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.914 |
-0.041 |
| 11 |
250646_at |
AT5G06720
|
ATPA2, peroxidase 2, AtPRX53, PA2, peroxidase 2, PRX53, peroxidase 53 |
0.900 |
-0.253 |
| 12 |
254649_at |
AT4G18570
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.877 |
0.599 |
| 13 |
266168_at |
AT2G38870
|
[Serine protease inhibitor, potato inhibitor I-type family protein] |
0.876 |
0.087 |
| 14 |
262880_at |
AT1G64880
|
[Ribosomal protein S5 family protein] |
0.860 |
0.338 |
| 15 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
0.860 |
-1.138 |
| 16 |
245145_at |
AT2G45440
|
DHDPS2, dihydrodipicolinate synthase 2 |
0.853 |
0.783 |
| 17 |
256983_at |
AT3G13470
|
Cpn60beta2, chaperonin-60beta2 |
0.852 |
0.400 |
| 18 |
247607_at |
AT5G60960
|
PNM1, PPR protein localized to the nucleus and mitochondria 1 |
0.845 |
-0.144 |
| 19 |
261460_at |
AT1G07880
|
ATMPK13 |
0.841 |
0.197 |
| 20 |
254314_at |
AT4G22470
|
[protease inhibitor/seed storage/lipid transfer protein (LTP) family protein] |
0.841 |
-0.437 |
| 21 |
247440_at |
AT5G62680
|
AtNPF2.11, GTR2, GLUCOSINOLATE TRANSPORTER-2, NPF2.11, NRT1/ PTR family 2.11 |
0.838 |
0.042 |
| 22 |
258532_at |
AT3G06700
|
[Ribosomal L29e protein family] |
0.837 |
1.480 |
| 23 |
251486_at |
AT3G59540
|
[Ribosomal L38e protein family] |
0.835 |
0.032 |
| 24 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.831 |
-0.364 |
| 25 |
246228_at |
AT4G36430
|
[Peroxidase superfamily protein] |
0.821 |
0.307 |
| 26 |
253203_at |
AT4G34710
|
ADC2, arginine decarboxylase 2, ATADC2, SPE2 |
0.820 |
1.046 |
| 27 |
255080_at |
AT4G09030
|
AGP10, arabinogalactan protein 10, ATAGP10 |
0.781 |
1.167 |
| 28 |
251846_at |
AT3G54560
|
HTA11, histone H2A 11 |
0.780 |
0.809 |
| 29 |
264262_at |
AT1G09200
|
H3.1, histone 3.1 |
0.753 |
0.559 |
| 30 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
0.746 |
1.389 |
| 31 |
263212_at |
AT1G10510
|
emb2004, embryo defective 2004 |
0.746 |
0.246 |
| 32 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.736 |
-0.634 |
| 33 |
259757_at |
AT1G77510
|
ATPDI6, PROTEIN DISULFIDE ISOMERASE 6, ATPDIL1-2, PDI-like 1-2, PDI6, PROTEIN DISULFIDE ISOMERASE 6, PDIL1-2, PDI-like 1-2 |
0.728 |
0.250 |
| 34 |
265076_at |
AT1G55490
|
CPN60B, chaperonin 60 beta, Cpn60beta1, chaperonin-60beta1, LEN1, LESION INITIATION 1 |
0.724 |
1.147 |
| 35 |
266256_at |
AT2G27710
|
[60S acidic ribosomal protein family] |
0.716 |
1.147 |
| 36 |
252272_at |
AT3G49670
|
BAM2, BARELY ANY MERISTEM 2 |
0.706 |
0.213 |
| 37 |
264689_at |
AT1G09900
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.704 |
-0.498 |
| 38 |
265671_at |
AT2G32060
|
[Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein] |
0.704 |
1.367 |
| 39 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
0.699 |
0.543 |
| 40 |
259196_at |
AT3G03600
|
RPS2, ribosomal protein S2 |
0.698 |
0.032 |
| 41 |
251966_at |
AT3G53380
|
LecRK-VIII.1, L-type lectin receptor kinase VIII.1 |
0.686 |
-0.349 |
| 42 |
253949_at |
AT4G26780
|
AR192, MGE2, mitochondrial GrpE 2 |
0.684 |
-0.387 |
| 43 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
0.674 |
0.307 |
| 44 |
251476_at |
AT3G59670
|
unknown |
0.673 |
-0.418 |
| 45 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
0.671 |
-0.292 |
| 46 |
264438_at |
AT1G27400
|
[Ribosomal protein L22p/L17e family protein] |
0.659 |
1.302 |
| 47 |
253566_at |
AT4G31210
|
[DNA topoisomerase, type IA, core] |
0.655 |
0.168 |
| 48 |
247654_at |
AT5G59850
|
[Ribosomal protein S8 family protein] |
0.647 |
0.579 |
| 49 |
247032_at |
AT5G67240
|
SDN3, small RNA degrading nuclease 3 |
0.645 |
0.145 |
| 50 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.643 |
0.057 |
| 51 |
251029_at |
AT5G02050
|
[Mitochondrial glycoprotein family protein] |
0.642 |
0.137 |
| 52 |
247109_at |
AT5G65870
|
ATPSK5, phytosulfokine 5 precursor, PSK5, PSK5, phytosulfokine 5 precursor |
0.636 |
0.673 |
| 53 |
247268_at |
AT5G64080
|
AtXYP1, XYP1, xylogen protein 1 |
0.635 |
-0.124 |
| 54 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.635 |
-0.386 |
| 55 |
253666_at |
AT4G30270
|
MERI-5, MERISTEM 5, MERI5B, meristem-5, SEN4, SENESCENCE 4, XTH24, xyloglucan endotransglucosylase/hydrolase 24 |
-1.000 |
0.263 |
| 56 |
245076_at |
AT2G23170
|
GH3.3 |
-0.759 |
-0.456 |
| 57 |
253010_at |
AT4G37750
|
ANT, AINTEGUMENTA, CKC, CKC1, COMPLEMENTING A PROTEIN KINASE C MUTANT 1, DRG, DRAGON |
-0.644 |
-0.502 |
| 58 |
251509_at |
AT3G59010
|
PME35, pectin methylesterase 35, PME61, pectin methylesterase 61 |
-0.638 |
0.336 |
| 59 |
262122_at |
AT1G02790
|
PGA4, polygalacturonase 4 |
-0.610 |
-0.642 |
| 60 |
259996_at |
AT1G67910
|
unknown |
-0.603 |
0.032 |
| 61 |
262438_at |
AT1G47410
|
unknown |
-0.597 |
0.007 |
| 62 |
255246_at |
AT4G05640
|
unknown |
-0.597 |
-1.004 |
| 63 |
253501_at |
AT4G32010
|
HSI2-L1, HSL1, HSI2-like 1, VAL2, VP1/ABI3-LIKE 2 |
-0.556 |
-0.023 |
| 64 |
257615_at |
AT3G26510
|
[Octicosapeptide/Phox/Bem1p family protein] |
-0.530 |
0.250 |
| 65 |
258797_at |
AT3G04730
|
IAA16, indoleacetic acid-induced protein 16 |
-0.499 |
1.353 |
| 66 |
258672_at |
AT3G08570
|
[Phototropic-responsive NPH3 family protein] |
-0.492 |
0.286 |
| 67 |
253193_at |
AT4G35380
|
[SEC7-like guanine nucleotide exchange family protein] |
-0.490 |
-0.953 |
| 68 |
267305_at |
AT2G30070
|
ATKT1, potassium transporter 1, ATKT1P, ATKUP1, KT1, potassium transporter 1, KUP1, POTASSIUM UPTAKE TRANSPORTER 1 |
-0.489 |
0.924 |
| 69 |
256435_at |
AT3G11180
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.488 |
-1.329 |
| 70 |
260242_at |
AT1G63650
|
AtEGL3, ATMYC-2, EGL1, EGL3, ENHANCER OF GLABRA 3 |
-0.486 |
-0.632 |
| 71 |
253588_at |
AT4G30790
|
[INVOLVED IN: autophagy] |
-0.478 |
-0.147 |
| 72 |
264083_at |
AT2G31230
|
ATERF15, ethylene-responsive element binding factor 15, ERF15, ethylene-responsive element binding factor 15 |
-0.464 |
0.432 |
| 73 |
248801_at |
AT5G47370
|
HAT2 |
-0.463 |
-0.065 |
| 74 |
250240_at |
AT5G13590
|
unknown |
-0.457 |
-0.043 |
| 75 |
258503_at |
AT3G02500
|
unknown |
-0.456 |
-0.993 |
| 76 |
249447_at |
AT5G39400
|
ATPTEN1, PTEN1, Phosphatase and TENsin homolog deleted on chromosome ten 1 |
-0.456 |
-0.937 |
| 77 |
261453_at |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
-0.454 |
0.365 |
| 78 |
262700_at |
AT1G76020
|
[Thioredoxin superfamily protein] |
-0.444 |
0.023 |
| 79 |
258726_at |
AT3G11745
|
unknown |
-0.444 |
-0.202 |
| 80 |
266111_at |
AT2G02061
|
[Nucleotide-diphospho-sugar transferase family protein] |
-0.442 |
-0.396 |
| 81 |
263942_at |
AT2G35860
|
FLA16, FASCICLIN-like arabinogalactan protein 16 precursor |
-0.440 |
0.306 |
| 82 |
262171_at |
AT1G74950
|
JAZ2, JASMONATE-ZIM-DOMAIN PROTEIN 2, TIFY10B |
-0.420 |
0.163 |
| 83 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
-0.420 |
0.377 |
| 84 |
264430_at |
AT1G61680
|
ATTPS14, TERPENE SYNTHASE 14, TPS14, terpene synthase 14 |
-0.407 |
-0.193 |
| 85 |
249306_at |
AT5G41400
|
[RING/U-box superfamily protein] |
-0.406 |
-0.102 |
| 86 |
256872_at |
AT3G26490
|
[Phototropic-responsive NPH3 family protein] |
-0.401 |
0.634 |
| 87 |
253038_at |
AT4G37790
|
HAT22 |
-0.380 |
0.601 |
| 88 |
266423_at |
AT2G41340
|
RPB5D, RNA polymerase II fifth largest subunit, D |
-0.378 |
-0.888 |
| 89 |
253966_at |
AT4G26520
|
AtFBA7, FBA7, fructose-bisphosphate aldolase 7 |
-0.377 |
-0.069 |
| 90 |
262228_at |
AT1G68690
|
AtPERK9, proline-rich extensin-like receptor kinase 9, PERK9, proline-rich extensin-like receptor kinase 9 |
-0.377 |
-0.025 |
| 91 |
245416_at |
AT4G17350
|
unknown |
-0.375 |
0.208 |
| 92 |
264826_at |
AT1G03410
|
2A6 |
-0.372 |
-0.663 |
| 93 |
262031_x_at |
AT1G37160
|
[gypsy-like retrotransposon family (Athila), similar to putative Athila retroelement ORF1 protein GI:4567296 from (Arabidopsis thaliana)] |
-0.368 |
-0.303 |
| 94 |
259535_at |
AT1G12280
|
SUMM2, suppressor of mkk1 mkk2 2 |
-0.364 |
-0.225 |
| 95 |
250216_at |
AT5G14090
|
AtLAZY1, LAZY1, LAZY 1 |
-0.353 |
0.243 |
| 96 |
251448_at |
AT3G59845
|
[Zinc-binding dehydrogenase family protein] |
-0.350 |
-0.064 |
| 97 |
252200_at |
AT3G50280
|
[HXXXD-type acyl-transferase family protein] |
-0.346 |
-0.891 |
| 98 |
261922_at |
AT1G65890
|
AAE12, acyl activating enzyme 12 |
-0.344 |
-1.180 |
| 99 |
250365_at |
AT5G11410
|
[Protein kinase superfamily protein] |
-0.341 |
-0.288 |
| 100 |
259000_at |
AT3G01860
|
unknown |
-0.341 |
0.449 |
| 101 |
255854_at |
AT1G67050
|
unknown |
-0.340 |
0.267 |
| 102 |
246999_at |
AT5G67440
|
MEL2, MAB4/ENP/NPY1-LIKE 2, NPY3, NAKED PINS IN YUC MUTANTS 3 |
-0.337 |
-0.559 |
| 103 |
249093_at |
AT5G43880
|
TRM21, TON1 Recruiting Motif 21 |
-0.328 |
0.561 |
| 104 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
-0.327 |
-0.231 |
| 105 |
264828_at |
AT1G03380
|
ATATG18G, homolog of yeast autophagy 18 (ATG18) G, ATG18G, homolog of yeast autophagy 18 (ATG18) G |
-0.322 |
-0.061 |
| 106 |
249647_at |
AT5G37040
|
[F-box family protein] |
-0.320 |
-0.867 |
| 107 |
251497_at |
AT3G59060
|
PIF5, PHYTOCHROME-INTERACTING FACTOR 5, PIL6, phytochrome interacting factor 3-like 6 |
-0.313 |
0.381 |