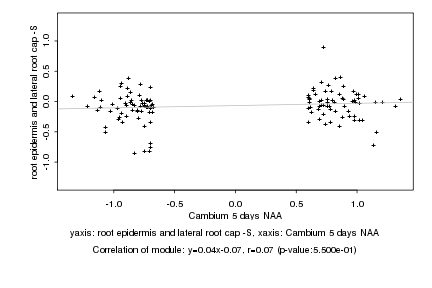
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Cambium 5 days NAA |
Log2 signal ratio
root epidermis and lateral root cap -S |
| 1 |
256825_at |
AT3G22120
|
CWLP, cell wall-plasma membrane linker protein |
1.354 |
0.044 |
| 2 |
250464_at |
AT5G10040
|
unknown |
1.316 |
-0.072 |
| 3 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
1.205 |
-0.008 |
| 4 |
263096_at |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
1.159 |
-0.509 |
| 5 |
261023_at |
AT1G12200
|
FMO, flavin monooxygenase |
1.151 |
-0.012 |
| 6 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
1.128 |
-0.720 |
| 7 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
1.053 |
0.083 |
| 8 |
259975_at |
AT1G76470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
1.041 |
-0.301 |
| 9 |
254726_at |
AT4G13660
|
ATPRR2, PRR2, pinoresinol reductase 2 |
1.012 |
-0.299 |
| 10 |
245385_at |
AT4G14020
|
[Rapid alkalinization factor (RALF) family protein] |
1.012 |
-0.017 |
| 11 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
1.011 |
0.052 |
| 12 |
252746_at |
AT3G43190
|
ATSUS4, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 4, SUS4, sucrose synthase 4 |
1.007 |
0.117 |
| 13 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.993 |
0.122 |
| 14 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
0.979 |
-0.005 |
| 15 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
0.978 |
-0.234 |
| 16 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
0.971 |
-0.299 |
| 17 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.971 |
0.031 |
| 18 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
0.969 |
0.174 |
| 19 |
258083_at |
AT3G25855
|
[Copper transport protein family] |
0.959 |
0.007 |
| 20 |
258064_at |
AT3G14680
|
CYP72A14, cytochrome P450, family 72, subfamily A, polypeptide 14 |
0.932 |
-0.231 |
| 21 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
0.927 |
-0.155 |
| 22 |
262325_at |
AT1G64160
|
AtDIR5, DIR5, dirigent protein 5 |
0.891 |
-0.078 |
| 23 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
0.885 |
0.251 |
| 24 |
248684_at |
AT5G48485
|
DIR1, DEFECTIVE IN INDUCED RESISTANCE 1 |
0.883 |
0.037 |
| 25 |
263680_at |
AT1G26930
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.878 |
-0.263 |
| 26 |
266141_at |
AT2G02120
|
LCR70, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 70, PDF2.1 |
0.877 |
0.066 |
| 27 |
245725_at |
AT1G73370
|
ATSUS6, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 6, SUS6, sucrose synthase 6 |
0.859 |
0.403 |
| 28 |
256646_at |
AT3G13590
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.850 |
0.130 |
| 29 |
254630_at |
AT4G18360
|
GOX3, glycolate oxidase 3 |
0.849 |
-0.407 |
| 30 |
250090_at |
AT5G17330
|
GAD, glutamate decarboxylase, GAD1, GLUTAMATE DECARBOXYLASE 1 |
0.817 |
-0.163 |
| 31 |
258969_at |
AT3G10680
|
[HSP20-like chaperones superfamily protein] |
0.817 |
0.384 |
| 32 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.811 |
-0.008 |
| 33 |
262912_at |
AT1G59740
|
AtNPF4.3, NPF4.3, NRT1/ PTR family 4.3 |
0.791 |
0.022 |
| 34 |
252457_at |
AT3G47180
|
[RING/U-box superfamily protein] |
0.788 |
0.181 |
| 35 |
252414_at |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
0.778 |
-0.330 |
| 36 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.775 |
-0.117 |
| 37 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.767 |
-0.077 |
| 38 |
250070_at |
AT5G17980
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
0.765 |
0.268 |
| 39 |
255111_at |
AT4G08780
|
[Peroxidase superfamily protein] |
0.755 |
-0.008 |
| 40 |
249531_at |
AT5G38770
|
AtGDU7, glutamine dumper 7, GDU7, glutamine dumper 7 |
0.753 |
0.037 |
| 41 |
247024_at |
AT5G66985
|
unknown |
0.752 |
-0.068 |
| 42 |
257654_at |
AT3G13310
|
DJC66, DNA J protein C66 |
0.752 |
-0.081 |
| 43 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
0.735 |
-0.367 |
| 44 |
266119_at |
AT2G02100
|
LCR69, low-molecular-weight cysteine-rich 69, PDF2.2 |
0.733 |
0.168 |
| 45 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
0.724 |
0.902 |
| 46 |
257153_at |
AT3G27220
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.721 |
-0.060 |
| 47 |
250152_at |
AT5G15120
|
unknown |
0.719 |
-0.199 |
| 48 |
254271_at |
AT4G23150
|
CRK7, cysteine-rich RLK (RECEPTOR-like protein kinase) 7 |
0.702 |
-0.056 |
| 49 |
260784_at |
AT1G06180
|
ATMYB13, myb domain protein 13, ATMYBLFGN, MYB13, myb domain protein 13 |
0.702 |
0.324 |
| 50 |
255080_at |
AT4G09030
|
AGP10, arabinogalactan protein 10, ATAGP10 |
0.702 |
0.017 |
| 51 |
262905_at |
AT1G59730
|
ATH7, thioredoxin H-type 7, TH7, thioredoxin H-type 7 |
0.685 |
0.006 |
| 52 |
265479_at |
AT2G15760
|
unknown |
0.684 |
-0.295 |
| 53 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
0.684 |
-0.081 |
| 54 |
259757_at |
AT1G77510
|
ATPDI6, PROTEIN DISULFIDE ISOMERASE 6, ATPDIL1-2, PDI-like 1-2, PDI6, PROTEIN DISULFIDE ISOMERASE 6, PDIL1-2, PDI-like 1-2 |
0.682 |
-0.123 |
| 55 |
252882_at |
AT4G39675
|
unknown |
0.655 |
0.126 |
| 56 |
267266_at |
AT2G23150
|
ATNRAMP3, NRAMP3, natural resistance-associated macrophage protein 3 |
0.637 |
0.182 |
| 57 |
261893_at |
AT1G80690
|
[PPPDE putative thiol peptidase family protein] |
0.634 |
0.227 |
| 58 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
0.622 |
-0.165 |
| 59 |
260528_at |
AT2G47260
|
ATWRKY23, WRKY DNA-BINDING PROTEIN 23, WRKY23, WRKY DNA-binding protein 23 |
0.614 |
-0.090 |
| 60 |
257952_at |
AT3G21770
|
[Peroxidase superfamily protein] |
0.607 |
0.054 |
| 61 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.606 |
-0.010 |
| 62 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
0.605 |
0.048 |
| 63 |
262001_at |
AT1G33790
|
[jacalin lectin family protein] |
0.600 |
0.113 |
| 64 |
259365_at |
AT1G13300
|
HRS1, HYPERSENSITIVITY TO LOW PI-ELICITED PRIMARY ROOT SHORTENING 1 |
0.599 |
-0.332 |
| 65 |
261930_at |
AT1G22440
|
[Zinc-binding alcohol dehydrogenase family protein] |
0.597 |
-0.104 |
| 66 |
245998_at |
AT5G20830
|
ASUS1, atsus1, SUS1, sucrose synthase 1 |
0.595 |
0.067 |
| 67 |
259788_at |
AT1G29670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-1.344 |
0.095 |
| 68 |
246380_at |
AT1G57750
|
CYP96A15, cytochrome P450, family 96, subfamily A, polypeptide 15, MAH1, MID-CHAIN ALKANE HYDROXYLASE 1 |
-1.219 |
-0.077 |
| 69 |
248793_at |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
-1.162 |
0.068 |
| 70 |
249227_at |
AT5G42180
|
PER64, peroxidase 64 |
-1.135 |
-0.133 |
| 71 |
264495_at |
AT1G27380
|
RIC2, ROP-interactive CRIB motif-containing protein 2 |
-1.123 |
0.178 |
| 72 |
256168_at |
AT1G51805
|
[Leucine-rich repeat protein kinase family protein] |
-1.113 |
-0.085 |
| 73 |
265894_at |
AT2G15050
|
LTP, lipid transfer protein, LTP7, lipid transfer protein 7 |
-1.108 |
0.021 |
| 74 |
263482_at |
AT2G03980
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-1.075 |
-0.504 |
| 75 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-1.073 |
-0.414 |
| 76 |
265342_at |
AT2G18300
|
HBI1, HOMOLOG OF BEE2 INTERACTING WITH IBH 1 |
-1.028 |
-0.151 |
| 77 |
254122_at |
AT4G24510
|
CER2, ECERIFERUM 2, VC-2, VC2 |
-1.016 |
-0.038 |
| 78 |
260267_at |
AT1G68530
|
CER6, ECERIFERUM 6, CUT1, CUTICULAR 1, G2, KCS6, 3-ketoacyl-CoA synthase 6, POP1, POLLEN-PISTIL INCOMPATIBILITY 1 |
-0.971 |
-0.099 |
| 79 |
247921_at |
AT5G57660
|
ATCOL5, BBX6, B-box domain protein 6, COL5, CONSTANS-like 5 |
-0.963 |
-0.282 |
| 80 |
255756_at |
AT1G19940
|
AtGH9B5, glycosyl hydrolase 9B5, GH9B5, glycosyl hydrolase 9B5 |
-0.959 |
-0.252 |
| 81 |
259466_at |
AT1G19050
|
ARR7, response regulator 7 |
-0.952 |
0.251 |
| 82 |
246275_at |
AT4G36540
|
BEE2, BR enhanced expression 2 |
-0.946 |
0.052 |
| 83 |
264774_at |
AT1G22890
|
unknown |
-0.944 |
-0.197 |
| 84 |
263709_at |
AT1G09310
|
unknown |
-0.939 |
0.311 |
| 85 |
267298_at |
AT2G23760
|
BLH4, BEL1-like homeodomain 4, SAW2, SAWTOOTH 2 |
-0.931 |
-0.342 |
| 86 |
261703_at |
AT1G32770
|
ANAC012, NAC domain containing protein 12, NAC012, NAC domain containing protein 12, NST3, NAC SECONDARY WALL THICKENING PROMOTING 3, SND1, SECONDARY WALL-ASSOCIATED NAC DOMAIN 1 |
-0.907 |
-0.025 |
| 87 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
-0.902 |
-0.244 |
| 88 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.901 |
-0.059 |
| 89 |
252280_at |
AT3G49260
|
iqd21, IQ-domain 21 |
-0.895 |
0.085 |
| 90 |
257757_at |
AT3G18660
|
GUX1, glucuronic acid substitution of xylan 1, PGSIP1, plant glycogenin-like starch initiation protein 1 |
-0.888 |
0.221 |
| 91 |
252543_at |
AT3G45780
|
JK224, NPH1, NONPHOTOTROPIC HYPOCOTYL 1, PHOT1, phototropin 1, RPT1, ROOT PHOTOTROPISM 1 |
-0.880 |
0.393 |
| 92 |
260547_at |
AT2G43550
|
[Scorpion toxin-like knottin superfamily protein] |
-0.869 |
0.159 |
| 93 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
-0.864 |
-0.001 |
| 94 |
257233_at |
AT3G15050
|
IQD10, IQ-domain 10 |
-0.857 |
0.017 |
| 95 |
251297_at |
AT3G62020
|
GLP10, germin-like protein 10 |
-0.851 |
-0.036 |
| 96 |
263841_at |
AT2G36870
|
AtXTH32, XTH32, xyloglucan endotransglucosylase/hydrolase 32 |
-0.849 |
-0.137 |
| 97 |
247882_at |
AT5G57785
|
unknown |
-0.837 |
-0.855 |
| 98 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
-0.836 |
-0.055 |
| 99 |
260326_at |
AT1G63910
|
AtMYB103, myb domain protein 103, MYB103, myb domain protein 103 |
-0.811 |
-0.163 |
| 100 |
263513_at |
AT2G12400
|
unknown |
-0.806 |
-0.146 |
| 101 |
260666_at |
AT1G19300
|
ATGATL1, GATL1, GALACTURONOSYLTRANSFERASE-LIKE 1, GLZ1, GAOLAOZHUANGREN 1, PARVUS, PARVUS |
-0.803 |
-0.271 |
| 102 |
261913_at |
AT1G65860
|
FMO GS-OX1, flavin-monooxygenase glucosinolate S-oxygenase 1 |
-0.793 |
0.102 |
| 103 |
262612_at |
AT1G14150
|
PnsL2, Photosynthetic NDH subcomplex L 2, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.787 |
-0.077 |
| 104 |
250182_at |
AT5G14470
|
AtGALK2, GALK2, galactokinase 2 |
-0.782 |
0.286 |
| 105 |
245970_at |
AT5G20710
|
BGAL7, beta-galactosidase 7 |
-0.776 |
-0.154 |
| 106 |
265511_at |
AT2G05540
|
[Glycine-rich protein family] |
-0.774 |
0.031 |
| 107 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.766 |
-0.017 |
| 108 |
257793_at |
AT3G26960
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.756 |
-0.078 |
| 109 |
252115_at |
AT3G51600
|
LTP5, lipid transfer protein 5 |
-0.754 |
-0.411 |
| 110 |
246908_at |
AT5G25610
|
ATRD22, RD22, RESPONSIVE TO DESSICATION 22 |
-0.752 |
-0.028 |
| 111 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
-0.749 |
-0.825 |
| 112 |
251303_at |
AT3G61910
|
ANAC066, NAC domain protein 66, NAC066, NAC domain protein 66, NST2, NAC SECONDARY WALL THICKENING PROMOTING FACTOR2 |
-0.748 |
-0.062 |
| 113 |
260205_at |
AT1G70700
|
JAZ9, JASMONATE-ZIM-DOMAIN PROTEIN 9, TIFY7 |
-0.738 |
0.022 |
| 114 |
251785_at |
AT3G55130
|
ABCG19, ATP-binding cassette G19, ATWBC19, white-brown complex homolog 19, WBC19, white-brown complex homolog 19 |
-0.734 |
-0.083 |
| 115 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
-0.728 |
0.025 |
| 116 |
252412_at |
AT3G47295
|
unknown |
-0.725 |
-0.068 |
| 117 |
267094_at |
AT2G38080
|
ATLMCO4, ARABIDOPSIS LACCASE-LIKE MULTICOPPER OXIDASE 4, IRX12, IRREGULAR XYLEM 12, LAC4, LACCASE 4, LMCO4, LACCASE-LIKE MULTICOPPER OXIDASE 4 |
-0.724 |
0.031 |
| 118 |
254998_at |
AT4G09760
|
[Protein kinase superfamily protein] |
-0.724 |
-0.095 |
| 119 |
257859_at |
AT3G12955
|
SAUR74, SMALL AUXIN UPREGULATED RNA 74 |
-0.712 |
-0.825 |
| 120 |
267141_at |
AT2G38090
|
[Duplicated homeodomain-like superfamily protein] |
-0.712 |
-0.168 |
| 121 |
257628_at |
AT3G26290
|
CYP71B26, cytochrome P450, family 71, subfamily B, polypeptide 26 |
-0.709 |
0.003 |
| 122 |
246097_at |
AT5G20270
|
HHP1, heptahelical transmembrane protein1 |
-0.706 |
-0.335 |
| 123 |
248121_at |
AT5G54690
|
GAUT12, galacturonosyltransferase 12, IRX8, IRREGULAR XYLEM 8, LGT6 |
-0.705 |
0.232 |
| 124 |
266526_at |
AT2G16980
|
[Major facilitator superfamily protein] |
-0.704 |
-0.687 |
| 125 |
260531_at |
AT2G47240
|
CER8, ECERIFERUM 8, LACS1, LONG-CHAIN ACYL-COA SYNTHASE 1 |
-0.704 |
-0.746 |
| 126 |
260395_at |
AT1G69780
|
ATHB13 |
-0.699 |
-0.101 |
| 127 |
249472_at |
AT5G39210
|
CRR7, CHLORORESPIRATORY REDUCTION 7 |
-0.699 |
0.027 |
| 128 |
260490_at |
AT1G51500
|
ABCG12, ATP-binding cassette G12, AtABCG12, ATWBC12, ARABIDOPSIS THALIANA WHITE-BROWN COMPLEX 12, CER5, ECERIFERUM 5, D3, WBC12, WHITE-BROWN COMPLEX 12 |
-0.692 |
-0.065 |
| 129 |
261928_at |
AT1G22480
|
[Cupredoxin superfamily protein] |
-0.686 |
-0.043 |
| 130 |
254924_at |
AT4G11330
|
ATMPK5, MAP kinase 5, MPK5, MAP kinase 5 |
-0.686 |
-0.178 |
| 131 |
264195_at |
AT1G22690
|
[Gibberellin-regulated family protein] |
-0.677 |
-0.092 |