|
probeID |
AGICode |
Annotation |
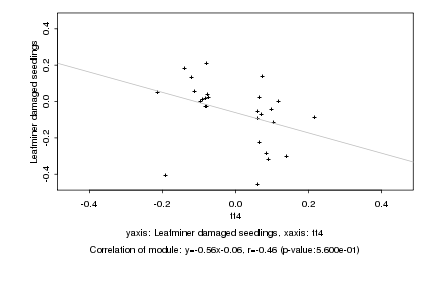
Log2 signal ratio
tt4 |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
246664_at |
AT5G34800
|
[pseudogene] |
0.215 |
-0.086 |
| 2 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
0.138 |
-0.297 |
| 3 |
251972_at |
AT3G53170
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.117 |
0.001 |
| 4 |
252223_at |
AT3G49850
|
ATTRB3, TBP2, TELOMERE-BINDING PROTEIN 2, TRB3, telomere repeat binding factor 3 |
0.102 |
-0.112 |
| 5 |
266824_at |
AT2G22800
|
HAT9 |
0.097 |
-0.042 |
| 6 |
260073_at |
AT1G73660
|
SIS8, SUGAR INSENSITIVE 8 |
0.088 |
-0.315 |
| 7 |
263252_at |
AT2G31380
|
BBX25, B-box domain protein 25, STH, salt tolerance homologue |
0.085 |
-0.284 |
| 8 |
256541_at |
AT1G42540
|
ATGLR3.3, GLR3.3, glutamate receptor 3.3 |
0.072 |
0.143 |
| 9 |
251404_at |
AT3G60310
|
unknown |
0.070 |
-0.071 |
| 10 |
260062_at |
AT1G73710
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.064 |
-0.220 |
| 11 |
256574_at |
AT3G14780
|
unknown |
0.064 |
0.024 |
| 12 |
245055_at |
AT2G26470
|
unknown |
0.060 |
-0.089 |
| 13 |
247100_at |
AT5G66520
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.059 |
-0.451 |
| 14 |
262565_at |
AT1G34320
|
unknown |
0.059 |
-0.054 |
| 15 |
260973_at |
AT1G53490
|
HEI10, homolog of human HEI10 ( Enhancer of cell Invasion No.10) |
-0.216 |
0.055 |
| 16 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
-0.193 |
-0.402 |
| 17 |
262782_at |
AT1G13195
|
[RING/U-box superfamily protein] |
-0.141 |
0.182 |
| 18 |
244921_s_at |
ATMG01000
|
ORF114 |
-0.122 |
0.135 |
| 19 |
257059_at |
AT3G15280
|
unknown |
-0.114 |
0.060 |
| 20 |
261163_x_at |
AT1G34410
|
ARF21, auxin response factor 21 |
-0.096 |
0.003 |
| 21 |
258745_at |
AT3G05920
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.091 |
0.015 |
| 22 |
257549_at |
AT3G18650
|
AGL103, AGAMOUS-like 103 |
-0.084 |
0.017 |
| 23 |
255658_at |
AT4G00770
|
TRM9, TON1 Recruiting Motif 9 |
-0.083 |
-0.022 |
| 24 |
253066_at |
AT4G37770
|
ACS8, 1-amino-cyclopropane-1-carboxylate synthase 8 |
-0.082 |
0.213 |
| 25 |
262855_at |
AT1G20860
|
PHT1;8, phosphate transporter 1;8 |
-0.080 |
-0.025 |
| 26 |
257945_at |
AT3G21860
|
ASK10, SKP1-like 10, SK10, SKP1-like 10 |
-0.079 |
0.043 |
| 27 |
256256_at |
AT3G11230
|
[Yippee family putative zinc-binding protein] |
-0.076 |
0.023 |