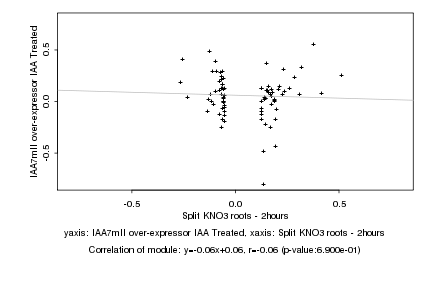
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Split KNO3 roots - 2hours |
Log2 signal ratio
IAA7mII over-expressor IAA Treated |
| 1 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
0.509 |
0.254 |
| 2 |
250472_at |
AT5G10210
|
unknown |
0.413 |
0.087 |
| 3 |
259365_at |
AT1G13300
|
HRS1, HYPERSENSITIVITY TO LOW PI-ELICITED PRIMARY ROOT SHORTENING 1 |
0.376 |
0.557 |
| 4 |
259681_at |
AT1G77760
|
GNR1, NIA1, nitrate reductase 1, NR1, NITRATE REDUCTASE 1 |
0.318 |
0.339 |
| 5 |
259813_at |
AT1G49860
|
ATGSTF14, glutathione S-transferase (class phi) 14, GSTF14, glutathione S-transferase (class phi) 14 |
0.307 |
0.070 |
| 6 |
257645_at |
AT3G25790
|
[myb-like transcription factor family protein] |
0.284 |
0.233 |
| 7 |
247443_at |
AT5G62720
|
[Integral membrane HPP family protein] |
0.258 |
0.133 |
| 8 |
261684_at |
AT1G47400
|
unknown |
0.235 |
0.103 |
| 9 |
265766_at |
AT2G48080
|
[oxidoreductase, 2OG-Fe(II) oxygenase family protein] |
0.232 |
0.312 |
| 10 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
0.223 |
0.076 |
| 11 |
246142_at |
AT5G19970
|
unknown |
0.211 |
0.155 |
| 12 |
262399_at |
AT1G49500
|
unknown |
0.206 |
0.121 |
| 13 |
246996_at |
AT5G67420
|
ASL39, ASYMMETRIC LEAVES2-LIKE 39, LBD37, LOB domain-containing protein 37 |
0.194 |
-0.068 |
| 14 |
253043_at |
AT4G37540
|
LBD39, LOB domain-containing protein 39 |
0.190 |
-0.434 |
| 15 |
265475_at |
AT2G15620
|
ATHNIR, ARABIDOPSIS THALIANA NITRITE REDUCTASE, NIR, NITRITE REDUCTASE, NIR1, nitrite reductase 1 |
0.189 |
-0.168 |
| 16 |
263319_at |
AT2G47160
|
AtBOR1, BOR1, REQUIRES HIGH BORON 1 |
0.188 |
0.014 |
| 17 |
260284_at |
AT1G80380
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.187 |
0.024 |
| 18 |
258629_at |
AT3G02850
|
SKOR, STELAR K+ outward rectifier |
0.187 |
0.001 |
| 19 |
255479_at |
AT4G02380
|
AtLEA5, Arabidopsis thaliana late embryogenensis abundant like 5, SAG21, senescence-associated gene 21 |
0.176 |
0.095 |
| 20 |
266313_at |
AT2G26980
|
CIPK3, CBL-interacting protein kinase 3, SnRK3.17, SNF1-RELATED PROTEIN KINASE 3.17 |
0.174 |
-0.024 |
| 21 |
245912_at |
AT5G19600
|
SULTR3;5, sulfate transporter 3;5 |
0.172 |
0.052 |
| 22 |
255734_at |
AT1G25550
|
[myb-like transcription factor family protein] |
0.170 |
0.125 |
| 23 |
264859_at |
AT1G24280
|
G6PD3, glucose-6-phosphate dehydrogenase 3 |
0.168 |
-0.249 |
| 24 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
0.166 |
0.069 |
| 25 |
262180_at |
AT1G78050
|
PGM, phosphoglycerate/bisphosphoglycerate mutase |
0.159 |
0.092 |
| 26 |
256788_at |
AT3G13730
|
CYP90D1, cytochrome P450, family 90, subfamily D, polypeptide 1 |
0.156 |
0.154 |
| 27 |
263096_at |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
0.150 |
0.113 |
| 28 |
250597_at |
AT5G07680
|
ANAC079, Arabidopsis NAC domain containing protein 79, ANAC080, NAC domain containing protein 80, ATNAC4, NAC080, NAC domain containing protein 80, NAC4 |
0.148 |
0.116 |
| 29 |
258437_at |
AT3G16560
|
[Protein phosphatase 2C family protein] |
0.147 |
0.371 |
| 30 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
0.143 |
-0.214 |
| 31 |
250463_at |
AT5G10030
|
OBF4, OCS ELEMENT BINDING FACTOR 4, TGA4, TGACG motif-binding factor 4 |
0.142 |
0.032 |
| 32 |
253049_at |
AT4G37300
|
MEE59, maternal effect embryo arrest 59 |
0.141 |
0.036 |
| 33 |
257467_at |
AT1G31320
|
LBD4, LOB domain-containing protein 4 |
0.138 |
0.043 |
| 34 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
0.138 |
0.020 |
| 35 |
253042_at |
AT4G37550
|
[Acetamidase/Formamidase family protein] |
0.132 |
-0.795 |
| 36 |
249927_at |
AT5G19220
|
ADG2, ADP GLUCOSE PYROPHOSPHORYLASE 2, APL1, ADP glucose pyrophosphorylase large subunit 1 |
0.131 |
-0.480 |
| 37 |
251090_at |
AT5G01340
|
AtmSFC1, mSFC1, mitochondrial succinate-fumarate carrier 1 |
0.125 |
-0.121 |
| 38 |
259466_at |
AT1G19050
|
ARR7, response regulator 7 |
0.125 |
0.128 |
| 39 |
254027_at |
AT4G25835
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.124 |
-0.058 |
| 40 |
256891_at |
AT3G19030
|
unknown |
0.124 |
-0.170 |
| 41 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
0.122 |
0.130 |
| 42 |
261806_at |
AT1G30510
|
ATRFNR2, root FNR 2, RFNR2, root FNR 2 |
0.121 |
-0.096 |
| 43 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
0.121 |
0.007 |
| 44 |
247591_at |
AT5G60770
|
ATNRT2.4, ARABIDOPSIS THALIANA NITRATE TRANSPORTER 2.4, NRT2.4, nitrate transporter 2.4 |
-0.270 |
0.190 |
| 45 |
264144_at |
AT1G79320
|
AtMC6, metacaspase 6, AtMCP2c, metacaspase 2c, MC6, metacaspase 6, MCP2c, metacaspase 2c |
-0.259 |
0.408 |
| 46 |
255957_at |
AT1G22160
|
unknown |
-0.234 |
0.048 |
| 47 |
250375_at |
AT5G11540
|
AtGulLO3, GulLO3, L -gulono-1,4-lactone ( L -GulL) oxidase 3 |
-0.137 |
-0.094 |
| 48 |
254037_at |
AT4G25760
|
ATGDU2, glutamine dumper 2, GDU2, glutamine dumper 2 |
-0.133 |
0.022 |
| 49 |
261956_at |
AT1G64590
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.130 |
0.493 |
| 50 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
-0.122 |
0.073 |
| 51 |
263828_at |
AT2G40250
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.120 |
0.009 |
| 52 |
253382_at |
AT4G33040
|
[Thioredoxin superfamily protein] |
-0.112 |
0.293 |
| 53 |
249522_at |
AT5G38700
|
unknown |
-0.109 |
-0.026 |
| 54 |
264323_at |
AT1G04180
|
YUC9, YUCCA 9 |
-0.101 |
0.100 |
| 55 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
-0.098 |
0.393 |
| 56 |
251689_at |
AT3G56500
|
[serine-rich protein-related] |
-0.093 |
0.299 |
| 57 |
254327_at |
AT4G22490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.080 |
0.198 |
| 58 |
248118_at |
AT5G55050
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.080 |
0.115 |
| 59 |
260540_at |
AT2G43500
|
NLP8, NIN-like protein 8 |
-0.077 |
-0.123 |
| 60 |
260945_at |
AT1G05950
|
unknown |
-0.076 |
0.290 |
| 61 |
252046_at |
AT3G52460
|
[hydroxyproline-rich glycoprotein family protein] |
-0.072 |
0.223 |
| 62 |
262236_at |
AT1G48330
|
unknown |
-0.072 |
0.244 |
| 63 |
262904_at |
AT1G59725
|
[DNAJ heat shock family protein] |
-0.071 |
0.073 |
| 64 |
267451_at |
AT2G33710
|
[Integrase-type DNA-binding superfamily protein] |
-0.071 |
0.131 |
| 65 |
262073_at |
AT1G59640
|
BPE, BIG PETAL, BPEp, BIG PETAL P, BPEub, BIG PETAL UB, ZCW32 |
-0.069 |
-0.244 |
| 66 |
264082_at |
AT2G28570
|
unknown |
-0.066 |
0.293 |
| 67 |
259805_at |
AT1G47890
|
AtRLP7, receptor like protein 7, RLP7, receptor like protein 7 |
-0.066 |
0.174 |
| 68 |
248734_at |
AT5G48090
|
ELP1, EDM2-like protein1 |
-0.065 |
-0.058 |
| 69 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
-0.064 |
0.128 |
| 70 |
258587_at |
AT3G04310
|
unknown |
-0.064 |
-0.174 |
| 71 |
258450_at |
AT3G22340
|
[copia-like retrotransposon family, has a 4.2e-60 P-value blast match to GB:AAC02666 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)] |
-0.063 |
0.167 |
| 72 |
256871_at |
AT3G26480
|
[Transducin family protein / WD-40 repeat family protein] |
-0.061 |
0.007 |
| 73 |
259827_at |
AT1G72270
|
unknown |
-0.061 |
-0.001 |
| 74 |
256609_at |
AT3G30843
|
unknown |
-0.061 |
0.001 |
| 75 |
252595_at |
AT3G45690
|
[Major facilitator superfamily protein] |
-0.060 |
0.042 |
| 76 |
249686_at |
AT5G36140
|
CYP716A2, cytochrome P450, family 716, subfamily A, polypeptide 2 |
-0.059 |
0.119 |
| 77 |
262722_at |
AT1G43620
|
TT15, TRANSPARENT TESTA 15, UGT80B1 |
-0.059 |
0.030 |
| 78 |
255837_at |
AT2G33460
|
RIC1, ROP-interactive CRIB motif-containing protein 1 |
-0.058 |
0.224 |
| 79 |
259722_at |
AT1G60930
|
ATRECQ4B, RECQ helicase L4B, RECQ4B, RECQ helicase L4B, RECQL4B, RECQ HELICASE L4B |
-0.057 |
-0.095 |
| 80 |
246075_at |
AT5G20410
|
ATMGD2, ARABIDOPSIS THALIANA MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE 2, MGD2, monogalactosyldiacylglycerol synthase 2 |
-0.057 |
-0.057 |
| 81 |
251594_at |
AT3G57630
|
[exostosin family protein] |
-0.056 |
0.128 |
| 82 |
263840_at |
AT2G36885
|
unknown |
-0.056 |
-0.191 |
| 83 |
262425_at |
AT1G47660
|
unknown |
-0.056 |
0.064 |
| 84 |
257142_at |
AT3G20090
|
CYP705A18, cytochrome P450, family 705, subfamily A, polypeptide 18 |
-0.056 |
-0.128 |
| 85 |
263292_at |
AT2G10900
|
[expressed protein] |
-0.055 |
-0.036 |