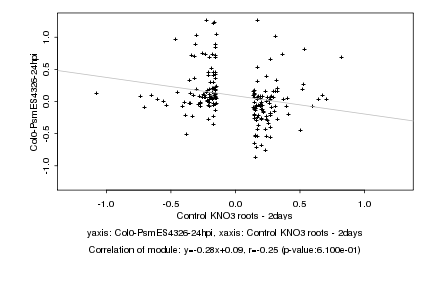
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Control KNO3 roots - 2days |
Log2 signal ratio
Col0-PsmES4326-24hpi |
| 1 |
252882_at |
AT4G39675
|
unknown |
0.821 |
0.686 |
| 2 |
262373_at |
AT1G73120
|
unknown |
0.704 |
0.035 |
| 3 |
253502_at |
AT4G31940
|
CYP82C4, cytochrome P450, family 82, subfamily C, polypeptide 4 |
0.669 |
0.099 |
| 4 |
254550_at |
AT4G19690
|
ATIRT1, ARABIDOPSIS IRON-REGULATED TRANSPORTER 1, IRT1, iron-regulated transporter 1 |
0.640 |
0.039 |
| 5 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
0.593 |
-0.071 |
| 6 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
0.529 |
0.811 |
| 7 |
259681_at |
AT1G77760
|
GNR1, NIA1, nitrate reductase 1, NR1, NITRATE REDUCTASE 1 |
0.526 |
0.266 |
| 8 |
258629_at |
AT3G02850
|
SKOR, STELAR K+ outward rectifier |
0.516 |
0.189 |
| 9 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
0.501 |
-0.443 |
| 10 |
255435_at |
AT4G03280
|
PETC, photosynthetic electron transfer C, PGR1, PROTON GRADIENT REGULATION 1 |
0.410 |
-0.201 |
| 11 |
249675_at |
AT5G35940
|
[Mannose-binding lectin superfamily protein] |
0.401 |
0.061 |
| 12 |
253317_at |
AT4G33960
|
unknown |
0.391 |
-0.065 |
| 13 |
262399_at |
AT1G49500
|
unknown |
0.370 |
0.041 |
| 14 |
252488_at |
AT3G46700
|
[UDP-Glycosyltransferase superfamily protein] |
0.360 |
0.736 |
| 15 |
256368_at |
AT1G66800
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.325 |
0.171 |
| 16 |
250472_at |
AT5G10210
|
unknown |
0.319 |
0.210 |
| 17 |
251664_at |
AT3G56940
|
ACSF, CHL27, CRD1, COPPER RESPONSE DEFECT 1 |
0.319 |
-0.267 |
| 18 |
250597_at |
AT5G07680
|
ANAC079, Arabidopsis NAC domain containing protein 79, ANAC080, NAC domain containing protein 80, ATNAC4, NAC080, NAC domain containing protein 80, NAC4 |
0.312 |
0.331 |
| 19 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
0.309 |
1.024 |
| 20 |
267526_at |
AT2G30570
|
PSBW, photosystem II reaction center W |
0.305 |
-0.068 |
| 21 |
262231_at |
AT1G68740
|
PHO1;H1 |
0.304 |
0.162 |
| 22 |
257807_at |
AT3G26650
|
GAPA, glyceraldehyde 3-phosphate dehydrogenase A subunit, GAPA-1, GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE A SUBUNIT 1 |
0.296 |
-0.143 |
| 23 |
249848_at |
AT5G23220
|
NIC3, nicotinamidase 3 |
0.293 |
0.158 |
| 24 |
251023_at |
AT5G02170
|
[Transmembrane amino acid transporter family protein] |
0.290 |
0.075 |
| 25 |
267612_at |
AT2G26690
|
AtNPF6.2, NPF6.2, NRT1/ PTR family 6.2 |
0.279 |
0.081 |
| 26 |
245044_at |
AT2G26500
|
[cytochrome b6f complex subunit (petM), putative] |
0.277 |
-0.079 |
| 27 |
264545_at |
AT1G55670
|
PSAG, photosystem I subunit G |
0.270 |
-0.172 |
| 28 |
254970_at |
AT4G10340
|
LHCB5, light harvesting complex of photosystem II 5 |
0.267 |
-0.185 |
| 29 |
266232_at |
AT2G02310
|
AtPP2-B6, phloem protein 2-B6, PP2-B6, phloem protein 2-B6 |
0.267 |
0.662 |
| 30 |
254213_at |
AT4G23590
|
[Tyrosine transaminase family protein] |
0.266 |
0.037 |
| 31 |
252858_at |
AT4G39770
|
TPPH, trehalose-6-phosphate phosphatase H |
0.266 |
-0.559 |
| 32 |
246142_at |
AT5G19970
|
unknown |
0.265 |
-0.204 |
| 33 |
259365_at |
AT1G13300
|
HRS1, HYPERSENSITIVITY TO LOW PI-ELICITED PRIMARY ROOT SHORTENING 1 |
0.264 |
0.067 |
| 34 |
261572_at |
AT1G01170
|
unknown |
0.264 |
-0.392 |
| 35 |
264594_at |
AT2G17640
|
ATSERAT3;1, SAT-106, SERINE ACETYLTRANSFERASE 106 |
0.262 |
-0.018 |
| 36 |
254828_at |
AT4G12550
|
AIR1, Auxin-Induced in Root cultures 1 |
0.261 |
0.029 |
| 37 |
251466_at |
AT3G59340
|
unknown |
0.257 |
0.027 |
| 38 |
254398_at |
AT4G21280
|
PSBQ, PHOTOSYSTEM II SUBUNIT Q, PSBQ-1, PHOTOSYSTEM II SUBUNIT Q-1, PSBQA, photosystem II subunit QA |
0.253 |
-0.328 |
| 39 |
259838_at |
AT1G52220
|
CURT1C, CURVATURE THYLAKOID 1C |
0.248 |
-0.291 |
| 40 |
260549_at |
AT2G43535
|
[Scorpion toxin-like knottin superfamily protein] |
0.244 |
0.063 |
| 41 |
262283_at |
AT1G68590
|
PSRP3/1, plastid-specific ribosomal protein 3/1 |
0.240 |
-0.539 |
| 42 |
251620_at |
AT3G58060
|
[Cation efflux family protein] |
0.240 |
0.009 |
| 43 |
264185_at |
AT1G54780
|
AtTLP18.3, TLP18.3, thylakoid lumen protein 18.3 |
0.237 |
-0.274 |
| 44 |
251545_at |
AT3G58810
|
ATMTP3, ARABIDOPSIS METAL TOLERANCE PROTEIN 3, ATMTPA2, MTP3, METAL TOLERANCE PROTEIN 3, MTPA2, metal tolerance protein A2 |
0.236 |
0.395 |
| 45 |
262134_at |
AT1G77990
|
AST56, SULTR2;2, SULPHATE TRANSPORTER 2;2 |
0.234 |
-0.220 |
| 46 |
259871_at |
AT1G76800
|
[Vacuolar iron transporter (VIT) family protein] |
0.231 |
-0.433 |
| 47 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
0.230 |
-0.749 |
| 48 |
252365_at |
AT3G48350
|
CEP3, cysteine endopeptidase 3 |
0.229 |
0.168 |
| 49 |
248151_at |
AT5G54270
|
LHCB3, light-harvesting chlorophyll B-binding protein 3, LHCB3*1 |
0.215 |
-0.160 |
| 50 |
255886_at |
AT1G20340
|
DRT112, DNA-DAMAGE-REPAIR/TOLERATION PROTEIN 112, PETE2, PLASTOCYANIN 2 |
0.213 |
-0.155 |
| 51 |
245061_at |
AT2G39730
|
RCA, rubisco activase |
0.209 |
-0.050 |
| 52 |
245912_at |
AT5G19600
|
SULTR3;5, sulfate transporter 3;5 |
0.204 |
0.079 |
| 53 |
252374_at |
AT3G48100
|
ARR5, response regulator 5, ATRR2, ARABIDOPSIS THALIANA RESPONSE REGULATOR 2, IBC6, INDUCED BY CYTOKININ 6, RR5, response regulator 5 |
0.202 |
-0.122 |
| 54 |
256244_at |
AT3G12520
|
SULTR4;2, sulfate transporter 4;2 |
0.201 |
-0.120 |
| 55 |
258087_at |
AT3G26060
|
ATPRX Q, PRXQ, peroxiredoxin Q |
0.198 |
-0.674 |
| 56 |
255290_at |
AT4G04640
|
ATPC1 |
0.197 |
-0.268 |
| 57 |
253043_at |
AT4G37540
|
LBD39, LOB domain-containing protein 39 |
0.196 |
-0.015 |
| 58 |
263319_at |
AT2G47160
|
AtBOR1, BOR1, REQUIRES HIGH BORON 1 |
0.196 |
-0.249 |
| 59 |
248728_at |
AT5G48000
|
CYP708 A2, CYTOCHROME P450, FAMILY 708, SUBFAMILY A, POLYPEPTIDE 2, CYP708A2, cytochrome P450, family 708, subfamily A, polypeptide 2, THAH, THALIANOL HYDROXYLASE, THAH1, THALIANOL HYDROXYLASE 1 |
0.194 |
0.080 |
| 60 |
263594_at |
AT2G01880
|
ATPAP7, PURPLE ACID PHOSPHATASE 7, PAP7, purple acid phosphatase 7 |
0.188 |
0.056 |
| 61 |
247483_at |
AT5G62420
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.187 |
-0.043 |
| 62 |
263805_at |
AT2G40400
|
unknown |
0.187 |
-0.069 |
| 63 |
253525_at |
AT4G31330
|
unknown |
0.175 |
0.082 |
| 64 |
261488_at |
AT1G14345
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.175 |
-0.370 |
| 65 |
247813_at |
AT5G58330
|
[lactate/malate dehydrogenase family protein] |
0.173 |
-0.203 |
| 66 |
259878_at |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
0.172 |
-0.053 |
| 67 |
259228_at |
AT3G07720
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.169 |
0.313 |
| 68 |
252183_at |
AT3G50740
|
UGT72E1, UDP-glucosyl transferase 72E1 |
0.169 |
-0.421 |
| 69 |
251293_at |
AT3G61930
|
unknown |
0.167 |
1.276 |
| 70 |
264246_at |
AT1G60140
|
ATTPS10, trehalose phosphate synthase, TPS10, trehalose phosphate synthase, TPS10, TREHALOSE PHOSPHATE SYNTHASE 10 |
0.165 |
0.541 |
| 71 |
248723_at |
AT5G47950
|
[HXXXD-type acyl-transferase family protein] |
0.165 |
0.071 |
| 72 |
261878_at |
AT1G50560
|
CYP705A25, cytochrome P450, family 705, subfamily A, polypeptide 25 |
0.164 |
0.046 |
| 73 |
259840_at |
AT1G52230
|
PSAH-2, PHOTOSYSTEM I SUBUNIT H-2, PSAH2, photosystem I subunit H2, PSI-H |
0.164 |
-0.245 |
| 74 |
250856_at |
AT5G04810
|
[pentatricopeptide (PPR) repeat-containing protein] |
0.164 |
-0.544 |
| 75 |
255302_at |
AT4G04830
|
ATMSRB5, methionine sulfoxide reductase B5, MSRB5, methionine sulfoxide reductase B5 |
0.160 |
-0.707 |
| 76 |
264052_at |
AT2G22330
|
CYP79B3, cytochrome P450, family 79, subfamily B, polypeptide 3 |
0.160 |
-0.058 |
| 77 |
247131_at |
AT5G66190
|
ATLFNR1, LEAF FNR 1, FNR1, ferredoxin-NADP(+)-oxidoreductase 1, LFNR1, leaf-type chloroplast-targeted FNR 1 |
0.159 |
-0.293 |
| 78 |
265033_at |
AT1G61520
|
LHCA3, photosystem I light harvesting complex gene 3 |
0.156 |
-0.076 |
| 79 |
253028_at |
AT4G38160
|
pde191, pigment defective 191 |
0.154 |
-0.520 |
| 80 |
260059_at |
AT1G78090
|
ATTPPB, Arabidopsis thaliana trehalose-6-phosphate phosphatase B, TPPB, trehalose-6-phosphate phosphatase B |
0.153 |
-0.098 |
| 81 |
256759_at |
AT3G25640
|
unknown |
0.153 |
-0.105 |
| 82 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
0.152 |
-0.861 |
| 83 |
251243_at |
AT3G61870
|
unknown |
0.151 |
-0.544 |
| 84 |
265374_at |
AT2G06520
|
PSBX, photosystem II subunit X |
0.150 |
-0.133 |
| 85 |
249927_at |
AT5G19220
|
ADG2, ADP GLUCOSE PYROPHOSPHORYLASE 2, APL1, ADP glucose pyrophosphorylase large subunit 1 |
0.147 |
-0.207 |
| 86 |
249535_at |
AT5G38820
|
[Transmembrane amino acid transporter family protein] |
0.146 |
-0.003 |
| 87 |
261746_at |
AT1G08380
|
PSAO, photosystem I subunit O |
0.146 |
-0.190 |
| 88 |
260812_at |
AT1G43650
|
UMAMIT22, Usually multiple acids move in and out Transporters 22 |
0.145 |
-0.249 |
| 89 |
252032_at |
AT3G52150
|
PSRP2, plastid-specific ribosomal protein 2 |
0.144 |
-0.643 |
| 90 |
265117_at |
AT1G62500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.144 |
0.120 |
| 91 |
256309_at |
AT1G30380
|
PSAK, photosystem I subunit K |
0.143 |
-0.101 |
| 92 |
245258_at |
AT4G15340
|
04C11, ATPEN1, pentacyclic triterpene synthase 1, PEN1, pentacyclic triterpene synthase 1 |
0.142 |
0.127 |
| 93 |
262412_at |
AT1G34760
|
GF14 OMICRON, GRF11, general regulatory factor 11, RHS5, ROOT HAIR SPECIFIC 5 |
0.142 |
-0.109 |
| 94 |
247436_at |
AT5G62530
|
ALDH12A1, aldehyde dehydrogenase 12A1, ATP5CDH, ARABIDOPSIS THALIANA DELTA1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE, P5CDH, DELTA1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE |
0.141 |
0.069 |
| 95 |
262133_at |
AT1G78000
|
SEL1, SELENATE RESISTANT 1, SULTR1;2, sulfate transporter 1;2 |
0.140 |
0.187 |
| 96 |
261177_at |
AT1G04770
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.139 |
-0.091 |
| 97 |
246825_at |
AT5G26260
|
[TRAF-like family protein] |
0.139 |
0.159 |
| 98 |
252920_at |
AT4G39000
|
AtGH9B17, glycosyl hydrolase 9B17, GH9B17, glycosyl hydrolase 9B17 |
0.138 |
0.094 |
| 99 |
253301_at |
AT4G33720
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-1.079 |
0.130 |
| 100 |
247591_at |
AT5G60770
|
ATNRT2.4, ARABIDOPSIS THALIANA NITRATE TRANSPORTER 2.4, NRT2.4, nitrate transporter 2.4 |
-0.739 |
0.086 |
| 101 |
249687_at |
AT5G36150
|
ATPEN3, putative pentacyclic triterpene synthase 3, PEN3, putative pentacyclic triterpene synthase 3 |
-0.710 |
-0.080 |
| 102 |
260851_at |
AT1G21890
|
UMAMIT19, Usually multiple acids move in and out Transporters 19 |
-0.651 |
0.105 |
| 103 |
260624_at |
AT1G08100
|
ACH2, ATNRT2.2, NITRATE TRANSPORTER 2.2, NRT2.2, nitrate transporter 2.2, NRT2;2AT |
-0.606 |
0.040 |
| 104 |
253036_at |
AT4G38340
|
NLP3, NIN-like protein 3 |
-0.564 |
0.014 |
| 105 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
-0.539 |
-0.049 |
| 106 |
256593_at |
AT3G28510
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.467 |
0.971 |
| 107 |
260298_at |
AT1G80320
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.452 |
0.155 |
| 108 |
253408_at |
AT4G32950
|
[Protein phosphatase 2C family protein] |
-0.417 |
-0.063 |
| 109 |
257668_at |
AT3G20460
|
[Major facilitator superfamily protein] |
-0.398 |
-0.006 |
| 110 |
250344_at |
AT5G11930
|
[Thioredoxin superfamily protein] |
-0.387 |
-0.209 |
| 111 |
255957_at |
AT1G22160
|
unknown |
-0.384 |
-0.511 |
| 112 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
-0.363 |
0.335 |
| 113 |
261574_at |
AT1G01190
|
CYP78A8, cytochrome P450, family 78, subfamily A, polypeptide 8 |
-0.358 |
-0.016 |
| 114 |
260540_at |
AT2G43500
|
NLP8, NIN-like protein 8 |
-0.356 |
-0.021 |
| 115 |
260030_at |
AT1G68880
|
AtbZIP, basic leucine-zipper 8, bZIP, basic leucine-zipper 8 |
-0.355 |
0.130 |
| 116 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
-0.342 |
0.720 |
| 117 |
258560_at |
AT3G06020
|
FAF4, FANTASTIC FOUR 4 |
-0.340 |
-0.219 |
| 118 |
263465_at |
AT2G31940
|
unknown |
-0.327 |
0.102 |
| 119 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
-0.324 |
0.701 |
| 120 |
253382_at |
AT4G33040
|
[Thioredoxin superfamily protein] |
-0.320 |
0.360 |
| 121 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
-0.317 |
0.901 |
| 122 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
-0.305 |
1.028 |
| 123 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
-0.303 |
0.200 |
| 124 |
247005_at |
AT5G67520
|
adenosine-5'-phosphosulfate (APS) kinase 4 |
-0.291 |
-0.045 |
| 125 |
245689_at |
AT5G04120
|
[Phosphoglycerate mutase family protein] |
-0.281 |
0.084 |
| 126 |
247212_at |
AT5G65040
|
unknown |
-0.278 |
-0.028 |
| 127 |
267298_at |
AT2G23760
|
BLH4, BEL1-like homeodomain 4, SAW2, SAWTOOTH 2 |
-0.276 |
-0.064 |
| 128 |
263828_at |
AT2G40250
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.274 |
-0.013 |
| 129 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
-0.260 |
0.758 |
| 130 |
253088_at |
AT4G36220
|
CYP84A1, CYTOCHROME P450 84A1, FAH1, ferulic acid 5-hydroxylase 1 |
-0.256 |
0.074 |
| 131 |
253182_at |
AT4G35190
|
LOG5, LONELY GUY 5 |
-0.254 |
0.123 |
| 132 |
264144_at |
AT1G79320
|
AtMC6, metacaspase 6, AtMCP2c, metacaspase 2c, MC6, metacaspase 6, MCP2c, metacaspase 2c |
-0.246 |
0.078 |
| 133 |
249686_at |
AT5G36140
|
CYP716A2, cytochrome P450, family 716, subfamily A, polypeptide 2 |
-0.244 |
0.105 |
| 134 |
261606_at |
AT1G49570
|
[Peroxidase superfamily protein] |
-0.241 |
0.095 |
| 135 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
-0.237 |
0.741 |
| 136 |
255893_at |
AT1G17960
|
[Threonyl-tRNA synthetase] |
-0.235 |
0.070 |
| 137 |
258570_at |
AT3G04530
|
ATPPCK2, PEPCK2, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 2, PPCK2, phosphoenolpyruvate carboxylase kinase 2 |
-0.231 |
0.147 |
| 138 |
259975_at |
AT1G76470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.229 |
1.263 |
| 139 |
249108_at |
AT5G43690
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.221 |
0.016 |
| 140 |
264083_at |
AT2G31230
|
ATERF15, ethylene-responsive element binding factor 15, ERF15, ethylene-responsive element binding factor 15 |
-0.221 |
0.014 |
| 141 |
260783_at |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
-0.219 |
0.175 |
| 142 |
248770_at |
AT5G47740
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.215 |
0.412 |
| 143 |
262244_at |
AT1G48260
|
CIPK17, CBL-interacting protein kinase 17, SnRK3.21, SNF1-RELATED PROTEIN KINASE 3.21 |
-0.214 |
0.455 |
| 144 |
249920_at |
AT5G19260
|
FAF3, FANTASTIC FOUR 3 |
-0.213 |
-0.276 |
| 145 |
258890_at |
AT3G05690
|
ATHAP2B, HEME ACTIVATOR PROTEIN (YEAST) HOMOLOG 2B, HAP2B, HEME ACTIVATOR PROTEIN (YEAST) HOMOLOG 2B, NF-YA2, nuclear factor Y, subunit A2, UNE8, UNFERTILIZED EMBRYO SAC 8 |
-0.210 |
-0.055 |
| 146 |
261991_at |
AT1G33700
|
[Beta-glucosidase, GBA2 type family protein] |
-0.207 |
-0.064 |
| 147 |
263572_at |
AT2G17060
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.206 |
-0.001 |
| 148 |
264562_at |
AT1G55760
|
[BTB/POZ domain-containing protein] |
-0.206 |
0.069 |
| 149 |
255923_at |
AT1G22180
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.205 |
0.700 |
| 150 |
261199_at |
AT1G12950
|
RSH2, root hair specific 2 |
-0.205 |
0.035 |
| 151 |
254797_at |
AT4G13030
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.203 |
0.121 |
| 152 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.202 |
0.189 |
| 153 |
263391_at |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
-0.197 |
0.066 |
| 154 |
260522_x_at |
AT2G41730
|
unknown |
-0.197 |
0.309 |
| 155 |
260599_at |
AT1G55940
|
CYP708A1, cytochrome P450, family 708, subfamily A, polypeptide 1 |
-0.192 |
0.068 |
| 156 |
247393_at |
AT5G63130
|
[Octicosapeptide/Phox/Bem1p family protein] |
-0.187 |
0.528 |
| 157 |
262378_at |
AT1G72830
|
ATHAP2C, HAP2C, NF-YA3, nuclear factor Y, subunit A3 |
-0.185 |
0.217 |
| 158 |
256081_at |
AT1G20700
|
ATWOX14, WUSCHEL RELATED HOMEOBOX 14, WOX14, WUSCHEL related homeobox 14 |
-0.185 |
0.051 |
| 159 |
256164_at |
AT1G48800
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.184 |
0.047 |
| 160 |
250100_at |
AT5G16570
|
GLN1;4, glutamine synthetase 1;4 |
-0.184 |
0.147 |
| 161 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
-0.178 |
0.742 |
| 162 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
-0.177 |
1.223 |
| 163 |
264901_at |
AT1G23090
|
AST91, sulfate transporter 91, SULTR3;3 |
-0.176 |
0.303 |
| 164 |
246597_at |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
-0.174 |
0.210 |
| 165 |
245401_at |
AT4G17670
|
unknown |
-0.174 |
0.405 |
| 166 |
257485_at |
AT1G63580
|
[Receptor-like protein kinase-related family protein] |
-0.173 |
-0.022 |
| 167 |
262811_at |
AT1G11700
|
unknown |
-0.171 |
-0.226 |
| 168 |
252501_at |
AT3G46880
|
unknown |
-0.171 |
-0.056 |
| 169 |
256011_at |
AT1G19230
|
[Riboflavin synthase-like superfamily protein] |
-0.171 |
0.464 |
| 170 |
247284_at |
AT5G64410
|
ATOPT4, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 4, OPT4, oligopeptide transporter 4 |
-0.171 |
-0.346 |
| 171 |
266965_at |
AT2G39510
|
UMAMIT14, Usually multiple acids move in and out Transporters 14 |
-0.169 |
0.094 |
| 172 |
265796_at |
AT2G35730
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.165 |
1.242 |
| 173 |
252340_at |
AT3G48920
|
AtMYB45, myb domain protein 45, MYB45, myb domain protein 45 |
-0.162 |
0.179 |
| 174 |
246616_at |
AT5G36260
|
[Eukaryotic aspartyl protease family protein] |
-0.162 |
0.116 |
| 175 |
257185_at |
AT3G13100
|
ABCC7, ATP-binding cassette C7, ATMRP7, multidrug resistance-associated protein 7, MRP7, MRP7, multidrug resistance-associated protein 7 |
-0.162 |
0.851 |
| 176 |
265016_at |
AT1G24420
|
[HXXXD-type acyl-transferase family protein] |
-0.161 |
0.051 |
| 177 |
259213_at |
AT3G09010
|
[Protein kinase superfamily protein] |
-0.161 |
0.365 |
| 178 |
251060_at |
AT5G01820
|
ATCIPK14, ATSR1, serine/threonine protein kinase 1, CIPK14, CBL-INTERACTING PROTEIN KINASE 14, PKS24, SOS2-like protein kinase 24, SnRK3.15, SNF1-RELATED PROTEIN KINASE 3.15, SR1, serine/threonine protein kinase 1 |
-0.161 |
0.195 |
| 179 |
249668_at |
AT5G35870
|
unknown |
-0.160 |
0.070 |
| 180 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
-0.160 |
0.087 |
| 181 |
265897_at |
AT2G25680
|
MOT1, molybdate transporter 1 |
-0.160 |
-0.036 |
| 182 |
247727_at |
AT5G59490
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.159 |
0.689 |
| 183 |
250098_at |
AT5G17350
|
unknown |
-0.159 |
0.224 |
| 184 |
264323_at |
AT1G04180
|
YUC9, YUCCA 9 |
-0.159 |
0.125 |
| 185 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.158 |
0.721 |
| 186 |
264082_at |
AT2G28570
|
unknown |
-0.157 |
0.455 |
| 187 |
248701_at |
AT5G48410
|
ATGLR1.3, ARABIDOPSIS THALIANA GLUTAMATE RECEPTOR 1.3, GLR1.3, glutamate receptor 1.3 |
-0.157 |
0.892 |
| 188 |
245510_at |
AT4G15740
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.157 |
0.063 |
| 189 |
246104_at |
AT5G28650
|
ATWRKY74, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 74, WRKY74, WRKY DNA-binding protein 74 |
-0.157 |
0.131 |
| 190 |
252205_at |
AT3G50350
|
unknown |
-0.156 |
-0.131 |
| 191 |
255283_at |
AT4G04620
|
ATG8B, autophagy 8b |
-0.155 |
0.425 |
| 192 |
252395_at |
AT3G47950
|
AHA4, H(+)-ATPase 4, HA4, H(+)-ATPase 4 |
-0.154 |
0.062 |
| 193 |
265723_at |
AT2G32140
|
[transmembrane receptors] |
-0.154 |
1.057 |
| 194 |
264759_at |
AT1G61480
|
[S-locus lectin protein kinase family protein] |
-0.152 |
0.242 |
| 195 |
253097_at |
AT4G37320
|
CYP81D5, cytochrome P450, family 81, subfamily D, polypeptide 5 |
-0.150 |
-0.019 |