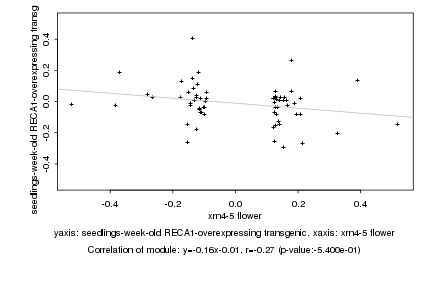
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
xrn4-5 flower |
Log2 signal ratio
seedlings-week-old RECA1-overexpressing transgenic |
| 1 |
248377_at |
AT5G51720
|
At-NEET, NEET, NEET group protein |
0.517 |
-0.143 |
| 2 |
251109_at |
AT5G01600
|
ATFER1, ARABIDOPSIS THALIANA FERRETIN 1, FER1, ferretin 1 |
0.388 |
0.137 |
| 3 |
258139_at |
AT3G24520
|
AT-HSFC1, HSFC1, heat shock transcription factor C1 |
0.324 |
-0.199 |
| 4 |
255645_at |
AT4G00880
|
SAUR31, SMALL AUXIN UPREGULATED RNA 31 |
0.213 |
-0.264 |
| 5 |
259704_at |
AT1G77680
|
[Ribonuclease II/R family protein] |
0.207 |
-0.077 |
| 6 |
246066_at |
AT5G19400
|
SMG7 |
0.207 |
0.025 |
| 7 |
260268_at |
AT1G68490
|
unknown |
0.193 |
-0.078 |
| 8 |
248592_at |
AT5G49280
|
[hydroxyproline-rich glycoprotein family protein] |
0.186 |
-0.008 |
| 9 |
259871_at |
AT1G76800
|
[Vacuolar iron transporter (VIT) family protein] |
0.177 |
0.070 |
| 10 |
248276_at |
AT5G53550
|
ATYSL3, YELLOW STRIPE LIKE 3, YSL3, YELLOW STRIPE like 3 |
0.177 |
0.266 |
| 11 |
256193_at |
AT1G30200
|
[F-box family protein] |
0.165 |
-0.021 |
| 12 |
253455_at |
AT4G32020
|
unknown |
0.160 |
0.008 |
| 13 |
257295_at |
AT3G17420
|
GPK1, glyoxysomal protein kinase 1 |
0.155 |
0.028 |
| 14 |
266770_at |
AT2G03090
|
ATEXP15, ATEXPA15, expansin A15, ATHEXP ALPHA 1.3, EXP15, EXPANSIN 15, EXPA15, expansin A15 |
0.152 |
-0.294 |
| 15 |
265291_at |
AT2G22720
|
[SPT2 chromatin protein] |
0.151 |
0.011 |
| 16 |
249261_at |
AT5G41710
|
[Mutator-like transposase family, has a 6.2e-18 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
0.141 |
0.031 |
| 17 |
258938_at |
AT3G10080
|
[RmlC-like cupins superfamily protein] |
0.138 |
-0.144 |
| 18 |
259984_at |
AT1G76460
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.138 |
0.010 |
| 19 |
253141_at |
AT4G35440
|
ATCLC-E, CLC-E, chloride channel E, CLCE, CHLORIDE CHANNEL E |
0.137 |
-0.126 |
| 20 |
261863_at |
AT1G50630
|
unknown |
0.134 |
-0.038 |
| 21 |
245078_at |
AT2G23340
|
DEAR3, DREB and EAR motif protein 3 |
0.130 |
-0.078 |
| 22 |
255626_at |
AT4G00780
|
[TRAF-like family protein] |
0.129 |
0.014 |
| 23 |
250424_at |
AT5G10550
|
GTE2, global transcription factor group E2 |
0.126 |
0.027 |
| 24 |
250372_at |
AT5G11460
|
unknown |
0.126 |
-0.150 |
| 25 |
248166_at |
AT5G54520
|
[Transducin/WD40 repeat-like superfamily protein] |
0.126 |
0.035 |
| 26 |
256976_at |
AT3G21020
|
[copia-like retrotransposon family, has a 5.4e-14 P-value blast match to gb|AAO73529.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
0.126 |
-0.035 |
| 27 |
254108_at |
AT4G25230
|
RIN2, RPM1 interacting protein 2 |
0.125 |
0.067 |
| 28 |
247735_at |
AT5G59440
|
ATTMPK.1, ARABIDOPSIS THALIANA THYMIDYLATE KINASE.1, ATTMPK.2, ARABIDOPSIS THALIANA THYMIDYLATE KINASE.2, ZEU1, ZEUS1 |
0.124 |
-0.067 |
| 29 |
245282_at |
AT4G14990
|
[Topoisomerase II-associated protein PAT1] |
0.123 |
-0.006 |
| 30 |
254632_at |
AT4G18630
|
unknown |
0.123 |
-0.251 |
| 31 |
257675_at |
AT3G20310
|
ATERF-7, ATERF7, ERF7, ethylene response factor 7 |
0.121 |
0.020 |
| 32 |
253270_at |
AT4G34160
|
CYCD3, CYCD3;1, CYCLIN D3;1 |
0.121 |
-0.166 |
| 33 |
267382_at |
AT2G44300
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.121 |
0.022 |
| 34 |
267295_at |
AT2G23800
|
GGPS2, geranylgeranyl pyrophosphate synthase 2, GGPS5, GERANYLGERANYL PYROPHOSPHATE SYNTHASE 5 |
-0.526 |
-0.019 |
| 35 |
260228_at |
AT1G74540
|
CYP98A8, cytochrome P450, family 98, subfamily A, polypeptide 8 |
-0.385 |
-0.024 |
| 36 |
262092_at |
AT1G56150
|
SAUR71, SMALL AUXIN UPREGULATED 71 |
-0.373 |
0.192 |
| 37 |
250904_at |
AT5G03620
|
[Subtilisin-like serine endopeptidase family protein] |
-0.281 |
0.046 |
| 38 |
260233_at |
AT1G74550
|
CYP98A9, cytochrome P450, family 98, subfamily A, polypeptide 9 |
-0.267 |
0.029 |
| 39 |
246592_at |
AT5G14890
|
[NHL domain-containing protein] |
-0.176 |
0.026 |
| 40 |
254211_at |
AT4G23570
|
SGT1A |
-0.174 |
0.132 |
| 41 |
266589_at |
AT2G46250
|
[myosin heavy chain-related] |
-0.156 |
-0.262 |
| 42 |
244937_at |
ATCG01110
|
NDHH, NAD(P)H dehydrogenase subunit H |
-0.155 |
-0.146 |
| 43 |
247567_at |
AT5G61190
|
[putative endonuclease or glycosyl hydrolase with C2H2-type zinc finger domain] |
-0.152 |
0.059 |
| 44 |
259719_at |
AT1G61070
|
LCR66, low-molecular-weight cysteine-rich 66, PDF2.4, PLANT DEFENSIN 2.4 |
-0.146 |
-0.021 |
| 45 |
250294_at |
AT5G13380
|
[Auxin-responsive GH3 family protein] |
-0.144 |
-0.010 |
| 46 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
-0.140 |
0.410 |
| 47 |
258140_at |
AT3G24503
|
ALDH1A, aldehyde dehydrogenase 1A, ALDH2C4, aldehyde dehydrogenase 2C4, REF1, REDUCED EPIDERMAL FLUORESCENCE1 |
-0.140 |
0.149 |
| 48 |
260143_at |
AT1G71880
|
ATSUC1, ARABIDOPSIS THALIANA SUCROSE-PROTON SYMPORTER 1, SUC1, sucrose-proton symporter 1 |
-0.135 |
0.087 |
| 49 |
245011_at |
ATCG00430
|
PSBG, photosystem II reaction center protein G |
-0.132 |
0.007 |
| 50 |
250635_at |
AT5G07510
|
ATGRP-4, ARABIDOPSIS THALIANA GLYCINE-RICH PROTEIN 4, ATGRP14, ARABIDOPSIS THALIANA GLYCINE-RICH PROTEIN 14, GRP-4, GLYCINE-RICH PROTEIN 4, GRP14, glycine-rich protein 14 |
-0.127 |
0.044 |
| 51 |
246963_at |
AT5G24820
|
[Eukaryotic aspartyl protease family protein] |
-0.126 |
0.031 |
| 52 |
266325_at |
AT2G46630
|
unknown |
-0.125 |
-0.175 |
| 53 |
254459_at |
AT4G21200
|
ATGA2OX8, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 8, GA2OX8, gibberellin 2-oxidase 8 |
-0.124 |
0.110 |
| 54 |
266965_at |
AT2G39510
|
UMAMIT14, Usually multiple acids move in and out Transporters 14 |
-0.120 |
0.190 |
| 55 |
245246_at |
AT1G44224
|
[ECA1 gametogenesis related family protein] |
-0.118 |
-0.039 |
| 56 |
263195_at |
AT1G36150
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.118 |
-0.049 |
| 57 |
251455_at |
AT3G60100
|
CSY5, citrate synthase 5 |
-0.114 |
0.021 |
| 58 |
250281_at |
AT5G13240
|
[transcription regulators] |
-0.112 |
-0.065 |
| 59 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
-0.111 |
-0.065 |
| 60 |
256860_at |
AT3G23840
|
CER26-LIKE, CER26-LIKE |
-0.103 |
-0.033 |
| 61 |
251641_at |
AT3G57470
|
[Insulinase (Peptidase family M16) family protein] |
-0.101 |
-0.078 |
| 62 |
258623_at |
AT3G02790
|
MBS1, METHYLENE BLUE SENSITIVITY 1 |
-0.099 |
-0.036 |
| 63 |
249650_at |
AT5G37055
|
ATSWC6, SEF, SERRATED LEAVES AND EARLY FLOWERING |
-0.097 |
0.002 |
| 64 |
249029_at |
AT5G44870
|
LAZ5, LAZARUS 5, TTR1, tolerance to Tobacco ringspot virus 1 |
-0.094 |
0.062 |
| 65 |
265379_at |
AT2G18340
|
[late embryogenesis abundant domain-containing protein / LEA domain-containing protein] |
-0.093 |
0.023 |