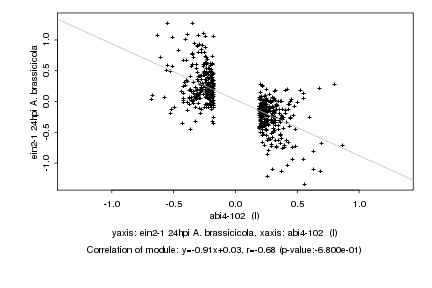
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
abi4-102 (I) |
Log2 signal ratio
ein2-1 24hpi A. brassicicola |
| 1 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
0.864 |
-0.707 |
| 2 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
0.794 |
0.282 |
| 3 |
252464_at |
AT3G47160
|
[RING/U-box superfamily protein] |
0.692 |
-0.672 |
| 4 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
0.687 |
-1.136 |
| 5 |
257365_x_at |
AT2G26020
|
PDF1.2b, plant defensin 1.2b |
0.674 |
0.225 |
| 6 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
0.629 |
-0.811 |
| 7 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
0.626 |
-1.098 |
| 8 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.596 |
-0.257 |
| 9 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
0.552 |
-1.332 |
| 10 |
260804_at |
AT1G78410
|
[VQ motif-containing protein] |
0.545 |
0.057 |
| 11 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
0.544 |
0.133 |
| 12 |
251673_at |
AT3G57240
|
AtBG3, BG3, beta-1,3-glucanase 3 |
0.544 |
-0.928 |
| 13 |
262421_at |
AT1G50290
|
unknown |
0.526 |
0.192 |
| 14 |
254566_at |
AT4G19240
|
unknown |
0.501 |
-0.011 |
| 15 |
256715_at |
AT2G34090
|
MEE18, maternal effect embryo arrest 18 |
0.480 |
-0.448 |
| 16 |
255607_at |
AT4G01130
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.479 |
-0.726 |
| 17 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
0.475 |
-0.079 |
| 18 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
0.462 |
-0.226 |
| 19 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
0.456 |
-0.940 |
| 20 |
250649_at |
AT5G06690
|
WCRKC1, WCRKC thioredoxin 1 |
0.454 |
-0.738 |
| 21 |
252068_at |
AT3G51440
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.448 |
0.040 |
| 22 |
265893_at |
AT2G15042
|
[Leucine-rich repeat (LRR) family protein] |
0.438 |
-0.274 |
| 23 |
252213_at |
AT3G50210
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.438 |
-0.252 |
| 24 |
263089_at |
AT2G16000
|
[copia-like retrotransposon family, has a 2.6e-248 P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
0.438 |
0.008 |
| 25 |
254252_at |
AT4G23310
|
CRK23, cysteine-rich RLK (RECEPTOR-like protein kinase) 23 |
0.434 |
-0.035 |
| 26 |
261613_at |
AT1G49720
|
ABF1, abscisic acid responsive element-binding factor 1, AtABF1 |
0.431 |
-0.402 |
| 27 |
246831_at |
AT5G26340
|
ATSTP13, SUGAR TRANSPORT PROTEIN 13, MSS1, STP13, SUGAR TRANSPORT PROTEIN 13 |
0.429 |
-0.123 |
| 28 |
255070_at |
AT4G09020
|
ATISA3, ISA3, isoamylase 3 |
0.427 |
-0.655 |
| 29 |
252098_at |
AT3G51330
|
[Eukaryotic aspartyl protease family protein] |
0.426 |
-0.041 |
| 30 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
0.420 |
0.201 |
| 31 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
0.419 |
-1.031 |
| 32 |
256455_at |
AT1G75190
|
unknown |
0.413 |
-0.338 |
| 33 |
262324_at |
AT1G64170
|
ATCHX16, cation/H+ exchanger 16, CHX16, cation/H+ exchanger 16 |
0.408 |
-0.623 |
| 34 |
265170_at |
AT1G23730
|
ATBCA3, BETA CARBONIC ANHYDRASE 3, BCA3, beta carbonic anhydrase 3 |
0.401 |
0.187 |
| 35 |
254093_at |
AT4G25110
|
AtMC2, metacaspase 2, AtMCP1c, metacaspase 1c, MC2, metacaspase 2, MCP1c, metacaspase 1c |
0.399 |
-0.181 |
| 36 |
262133_at |
AT1G78000
|
SEL1, SELENATE RESISTANT 1, SULTR1;2, sulfate transporter 1;2 |
0.397 |
-0.138 |
| 37 |
247216_at |
AT5G64860
|
DPE1, disproportionating enzyme |
0.390 |
-0.739 |
| 38 |
253981_at |
AT4G26670
|
[Mitochondrial import inner membrane translocase subunit Tim17/Tim22/Tim23 family protein] |
0.389 |
-0.708 |
| 39 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
0.388 |
-0.257 |
| 40 |
261937_at |
AT1G22570
|
[Major facilitator superfamily protein] |
0.387 |
-0.438 |
| 41 |
246929_at |
AT5G25210
|
unknown |
0.380 |
-0.756 |
| 42 |
262942_at |
AT1G79450
|
ALIS5, ALA-interacting subunit 5 |
0.380 |
-0.249 |
| 43 |
254716_at |
AT4G13560
|
UNE15, unfertilized embryo sac 15 |
0.377 |
-0.117 |
| 44 |
250302_at |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
0.374 |
-0.280 |
| 45 |
263367_at |
AT2G20460
|
[copia-like retrotransposon family, has a 0. P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
0.371 |
0.006 |
| 46 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
0.370 |
-0.598 |
| 47 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
0.369 |
-0.216 |
| 48 |
254350_at |
AT4G22280
|
[F-box/RNI-like superfamily protein] |
0.367 |
0.011 |
| 49 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.365 |
-1.131 |
| 50 |
257591_at |
AT3G24900
|
AtRLP39, receptor like protein 39, RLP39, receptor like protein 39 |
0.364 |
-0.268 |
| 51 |
252175_at |
AT3G50700
|
AtIDD2, indeterminate(ID)-domain 2, IDD2, indeterminate(ID)-domain 2 |
0.364 |
-0.531 |
| 52 |
266839_at |
AT2G25930
|
ELF3, EARLY FLOWERING 3, PYK20 |
0.362 |
-0.531 |
| 53 |
253331_at |
AT4G33490
|
[Eukaryotic aspartyl protease family protein] |
0.362 |
-0.667 |
| 54 |
257460_at |
AT1G75580
|
SAUR51, SMALL AUXIN UPREGULATED RNA 51 |
0.357 |
0.014 |
| 55 |
253679_at |
AT4G29610
|
[Cytidine/deoxycytidylate deaminase family protein] |
0.357 |
-0.536 |
| 56 |
248971_at |
AT5G45000
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.357 |
-0.339 |
| 57 |
259385_at |
AT1G13470
|
unknown |
0.356 |
-0.298 |
| 58 |
249843_at |
AT5G23570
|
ATSGS3, SUPPRESSOR OF GENE SILENCING 3, SGS3, SUPPRESSOR OF GENE SILENCING 3 |
0.354 |
-0.129 |
| 59 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
0.353 |
-0.184 |
| 60 |
259517_at |
AT1G20630
|
CAT1, catalase 1 |
0.347 |
0.009 |
| 61 |
263919_at |
AT2G36470
|
unknown |
0.346 |
-0.621 |
| 62 |
263128_at |
AT1G78600
|
BBX22, B-box domain protein 22, DBB3, DOUBLE B-BOX 3, LZF1, light-regulated zinc finger protein 1, STH3, SALT TOLERANCE HOMOLOG 3 |
0.346 |
-0.448 |
| 63 |
259992_at |
AT1G67970
|
AT-HSFA8, HSFA8, heat shock transcription factor A8 |
0.342 |
-0.374 |
| 64 |
259925_at |
AT1G75040
|
PR-5, PR5, pathogenesis-related gene 5 |
0.335 |
-0.059 |
| 65 |
260396_at |
AT1G69720
|
HO3, heme oxygenase 3 |
0.331 |
-0.075 |
| 66 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
0.331 |
-0.128 |
| 67 |
257466_at |
AT1G62840
|
unknown |
0.329 |
-0.038 |
| 68 |
261927_at |
AT1G22500
|
AtATL15, Arabidopsis thaliana Arabidopsis toxicos en levadura 15, ATL15, Arabidopsis toxicos en levadura 15 |
0.327 |
-0.061 |
| 69 |
246371_at |
AT1G51940
|
LYK3, LysM-containing receptor-like kinase 3 |
0.327 |
-0.718 |
| 70 |
263548_at |
AT2G21660
|
ATGRP7, GLYCINE RICH PROTEIN 7, CCR2, cold, circadian rhythm, and rna binding 2, GR-RBP7, GLYCINE-RICH RNA-BINDING PROTEIN 7, GRP7, GLYCINE-RICH RNA-BINDING PROTEIN 7 |
0.325 |
-0.734 |
| 71 |
248709_at |
AT5G48470
|
PRDA1, PEP-Related Development Arrested 1 |
0.324 |
-0.259 |
| 72 |
259293_at |
AT3G11580
|
[AP2/B3-like transcriptional factor family protein] |
0.323 |
0.194 |
| 73 |
246343_at |
AT3G56720
|
unknown |
0.323 |
-0.382 |
| 74 |
247685_at |
AT5G59680
|
[Leucine-rich repeat protein kinase family protein] |
0.320 |
-0.337 |
| 75 |
264045_at |
AT2G22450
|
AtRIBA2, RIBA2, homolog of ribA 2 |
0.319 |
-0.631 |
| 76 |
259928_at |
AT1G34380
|
[5'-3' exonuclease family protein] |
0.319 |
-0.326 |
| 77 |
263852_at |
AT2G04450
|
ATNUDT6, nudix hydrolase homolog 6, ATNUDX6, Arabidopsis thaliana nucleoside diphosphate linked to some moiety X 6, NUDT6, nudix hydrolase homolog 6, NUDX6, nucleoside diphosphates linked to some moiety X 6 |
0.318 |
0.030 |
| 78 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
0.318 |
-0.136 |
| 79 |
251631_at |
AT3G57430
|
OTP84, ORGANELLE TRANSCRIPT PROCESSING 84 |
0.318 |
-0.301 |
| 80 |
259211_at |
AT3G09020
|
[alpha 1,4-glycosyltransferase family protein] |
0.313 |
-0.185 |
| 81 |
247071_at |
AT5G66640
|
DAR3, DA1-related protein 3 |
0.312 |
-0.257 |
| 82 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
0.311 |
-0.295 |
| 83 |
255381_at |
AT4G03510
|
ATRMA1, RMA1, RING membrane-anchor 1 |
0.310 |
-0.461 |
| 84 |
256883_at |
AT3G26440
|
unknown |
0.309 |
0.166 |
| 85 |
252076_at |
AT3G51660
|
[Tautomerase/MIF superfamily protein] |
0.309 |
0.058 |
| 86 |
247225_at |
AT5G65090
|
BST1, BRISTLED 1, DER4, DEFORMED ROOT HAIRS 4, MRH3 |
0.309 |
-0.146 |
| 87 |
246238_at |
AT4G36670
|
AtPLT6, AtPMT6, PLT6, polyol transporter 6, PMT6, polyol/monosaccharide transporter 6 |
0.309 |
-0.170 |
| 88 |
251232_at |
AT3G62780
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.306 |
-0.484 |
| 89 |
254229_at |
AT4G23610
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.305 |
0.023 |
| 90 |
259757_at |
AT1G77510
|
ATPDI6, PROTEIN DISULFIDE ISOMERASE 6, ATPDIL1-2, PDI-like 1-2, PDI6, PROTEIN DISULFIDE ISOMERASE 6, PDIL1-2, PDI-like 1-2 |
0.305 |
-0.145 |
| 91 |
251789_at |
AT3G55450
|
PBL1, PBS1-like 1 |
0.305 |
-0.432 |
| 92 |
254645_at |
AT4G18520
|
SEL1, SEEDLING LETHAL 1 |
0.303 |
-0.502 |
| 93 |
259489_at |
AT1G15790
|
unknown |
0.303 |
-0.089 |
| 94 |
263133_at |
AT1G78450
|
[SOUL heme-binding family protein] |
0.303 |
-0.232 |
| 95 |
258791_at |
AT3G04720
|
AtPR4, HEL, HEVEIN-LIKE, PR-4, PR4, pathogenesis-related 4 |
0.302 |
0.062 |
| 96 |
261453_at |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
0.301 |
-0.331 |
| 97 |
258151_at |
AT3G18080
|
BGLU44, B-S glucosidase 44 |
0.301 |
-0.436 |
| 98 |
267509_at |
AT2G45660
|
AGL20, AGAMOUS-like 20, ATSOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1, SOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1 |
0.301 |
-0.258 |
| 99 |
245644_at |
AT1G25320
|
[Leucine-rich repeat protein kinase family protein] |
0.300 |
-0.343 |
| 100 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.299 |
-0.068 |
| 101 |
253085_s_at |
AT4G36280
|
CRH1, CRT1 Homologue 1, MORC2, microrchidia 2 |
0.299 |
-0.095 |
| 102 |
258191_at |
AT3G29100
|
ATVTI13, VTI13, vesicle transport V-snare 13 |
0.298 |
0.011 |
| 103 |
245932_at |
AT5G09290
|
[Inositol monophosphatase family protein] |
0.298 |
0.073 |
| 104 |
262772_at |
AT1G13210
|
ACA.l, autoinhibited Ca2+/ATPase II |
0.298 |
-0.314 |
| 105 |
251919_at |
AT3G53800
|
Fes1B, Fes1B |
0.298 |
-0.572 |
| 106 |
266761_at |
AT2G47130
|
AtSDR3, SDR3, short-chain dehydrogenase/reductase 2 |
0.295 |
-0.192 |
| 107 |
253581_at |
AT4G30660
|
[Low temperature and salt responsive protein family] |
0.292 |
-1.090 |
| 108 |
253140_at |
AT4G35480
|
RHA3B, RING-H2 finger A3B |
0.291 |
-0.301 |
| 109 |
253626_at |
AT4G30640
|
[RNI-like superfamily protein] |
0.291 |
-0.276 |
| 110 |
245188_at |
AT1G67660
|
[Restriction endonuclease, type II-like superfamily protein] |
0.289 |
-0.506 |
| 111 |
248092_at |
AT5G55170
|
ATSUMO3, SUM3, SMALL UBIQUITIN-LIKE MODIFIER 3, SUMO 3, SMALL UBIQUITIN-LIKE MODIFIER 3, SUMO3, small ubiquitin-like modifier 3 |
0.286 |
-0.226 |
| 112 |
255232_at |
AT4G05330
|
AGD13, ARF-GAP domain 13 |
0.285 |
-0.336 |
| 113 |
247604_at |
AT5G60950
|
COBL5, COBRA-like protein 5 precursor |
0.285 |
-0.015 |
| 114 |
267355_at |
AT2G39900
|
WLIM2a, WLIM2a |
0.285 |
-0.682 |
| 115 |
248895_at |
AT5G46330
|
FLS2, FLAGELLIN-SENSITIVE 2 |
0.284 |
-0.730 |
| 116 |
264246_at |
AT1G60140
|
ATTPS10, trehalose phosphate synthase, TPS10, trehalose phosphate synthase, TPS10, TREHALOSE PHOSPHATE SYNTHASE 10 |
0.283 |
0.031 |
| 117 |
245353_at |
AT4G16000
|
unknown |
0.283 |
-0.110 |
| 118 |
249346_at |
AT5G40780
|
LHT1, lysine histidine transporter 1 |
0.282 |
-0.106 |
| 119 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
0.280 |
-0.146 |
| 120 |
254011_at |
AT4G26370
|
[antitermination NusB domain-containing protein] |
0.279 |
-0.606 |
| 121 |
249397_at |
AT5G40230
|
UMAMIT37, Usually multiple acids move in and out Transporters 37 |
0.276 |
0.000 |
| 122 |
252698_at |
AT3G43670
|
[Copper amine oxidase family protein] |
0.275 |
0.119 |
| 123 |
254931_at |
AT4G11460
|
CRK30, cysteine-rich RLK (RECEPTOR-like protein kinase) 30 |
0.274 |
-0.071 |
| 124 |
255941_at |
AT1G20350
|
ATTIM17-1, translocase inner membrane subunit 17-1, TIM17-1, translocase inner membrane subunit 17-1 |
0.273 |
-0.095 |
| 125 |
255500_at |
AT4G02390
|
APP, poly(ADP-ribose) polymerase, PARP2, poly(ADP-ribose) polymerase 2, PP, poly(ADP-ribose) polymerase |
0.272 |
-0.113 |
| 126 |
253871_at |
AT4G27440
|
PORB, protochlorophyllide oxidoreductase B |
0.271 |
-0.603 |
| 127 |
266681_at |
AT2G19840
|
[copia-like retrotransposon family, has a 3.5e-301 P-value blast match to GB:CAA31653 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)] |
0.266 |
0.001 |
| 128 |
251221_at |
AT3G62550
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.266 |
-0.788 |
| 129 |
247786_at |
AT5G58600
|
PMR5, POWDERY MILDEW RESISTANT 5, TBL44, TRICHOME BIREFRINGENCE-LIKE 44 |
0.264 |
-0.585 |
| 130 |
255485_at |
AT4G02550
|
unknown |
0.264 |
-0.228 |
| 131 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
0.262 |
-0.160 |
| 132 |
261125_at |
AT1G04990
|
[Zinc finger C-x8-C-x5-C-x3-H type family protein] |
0.262 |
-0.296 |
| 133 |
266016_at |
AT2G18670
|
[RING/U-box superfamily protein] |
0.261 |
-0.136 |
| 134 |
259441_at |
AT1G02300
|
[Cysteine proteinases superfamily protein] |
0.260 |
-0.370 |
| 135 |
251346_at |
AT3G60980
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.260 |
-0.132 |
| 136 |
266668_at |
AT2G29760
|
OTP81, ORGANELLE TRANSCRIPT PROCESSING 81 |
0.260 |
-0.376 |
| 137 |
250987_at |
AT5G02860
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.258 |
-0.342 |
| 138 |
247348_at |
AT5G63810
|
AtBGAL10, Arabidopsis thaliana beta-galactosidase 10, BGAL10, beta-galactosidase 10 |
0.258 |
-0.849 |
| 139 |
256746_at |
AT3G29320
|
PHS1, alpha-glucan phosphorylase 1 |
0.257 |
-0.619 |
| 140 |
266797_at |
AT2G22840
|
AtGRF1, growth-regulating factor 1, GRF1, growth-regulating factor 1 |
0.257 |
-0.289 |
| 141 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
0.256 |
-1.213 |
| 142 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
0.255 |
0.024 |
| 143 |
261167_at |
AT1G04980
|
ATPDI10, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 10, ATPDIL2-2, PDI-like 2-2, PDI10, PROTEIN DISULFIDE ISOMERASE, PDIL2-2, PDI-like 2-2 |
0.254 |
-0.166 |
| 144 |
261254_at |
AT1G05805
|
AKS2, ABA-responsive kinase substrate 2 |
0.250 |
-0.276 |
| 145 |
259902_at |
AT1G74170
|
AtRLP13, receptor like protein 13, RLP13, receptor like protein 13 |
0.249 |
-0.214 |
| 146 |
254922_at |
AT4G11370
|
RHA1A, RING-H2 finger A1A |
0.249 |
-0.310 |
| 147 |
266658_at |
AT2G25735
|
unknown |
0.247 |
-0.076 |
| 148 |
257925_at |
AT3G23170
|
unknown |
0.246 |
-0.118 |
| 149 |
253421_at |
AT4G32340
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.246 |
-0.470 |
| 150 |
250127_at |
AT5G16380
|
unknown |
0.246 |
-0.444 |
| 151 |
264574_at |
AT1G05300
|
ZIP5, zinc transporter 5 precursor |
0.246 |
-0.539 |
| 152 |
260146_at |
AT1G52770
|
[Phototropic-responsive NPH3 family protein] |
0.245 |
0.057 |
| 153 |
255481_at |
AT4G02460
|
AtPMS1, PMS1, POSTMEIOTIC SEGREGATION 1 |
0.245 |
0.001 |
| 154 |
251347_at |
AT3G61010
|
[Ferritin/ribonucleotide reductase-like family protein] |
0.244 |
-0.038 |
| 155 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
0.244 |
0.197 |
| 156 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
0.244 |
-0.165 |
| 157 |
260728_at |
AT1G48210
|
[Protein kinase superfamily protein] |
0.243 |
-0.286 |
| 158 |
248820_at |
AT5G47060
|
unknown |
0.243 |
0.025 |
| 159 |
246881_at |
AT5G26040
|
HDA2, histone deacetylase 2 |
0.242 |
-0.347 |
| 160 |
262881_at |
AT1G64890
|
[Major facilitator superfamily protein] |
0.242 |
-0.441 |
| 161 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.242 |
-0.045 |
| 162 |
263438_at |
AT2G28660
|
[Chloroplast-targeted copper chaperone protein] |
0.241 |
-0.031 |
| 163 |
259735_at |
AT1G64405
|
unknown |
0.240 |
0.038 |
| 164 |
262641_at |
AT1G62730
|
[Terpenoid synthases superfamily protein] |
0.240 |
-0.120 |
| 165 |
246855_at |
AT5G26280
|
[TRAF-like family protein] |
0.239 |
0.104 |
| 166 |
248914_at |
AT5G45750
|
AtRABA1c, RAB GTPase homolog A1C, RABA1c, RAB GTPase homolog A1C |
0.239 |
-0.315 |
| 167 |
264314_at |
AT1G70420
|
unknown |
0.239 |
-0.040 |
| 168 |
263182_at |
AT1G05575
|
unknown |
0.237 |
-0.080 |
| 169 |
250381_at |
AT5G11610
|
[Exostosin family protein] |
0.237 |
-0.210 |
| 170 |
246082_at |
AT5G20480
|
EFR, EF-TU receptor |
0.237 |
-0.091 |
| 171 |
265790_at |
AT2G01170
|
AtGABP, BAT1, bidirectional amino acid transporter 1, GABP, GABA permease |
0.236 |
-0.387 |
| 172 |
252053_at |
AT3G52400
|
ATSYP122, SYP122, syntaxin of plants 122 |
0.236 |
-0.058 |
| 173 |
247800_at |
AT5G58570
|
unknown |
0.234 |
-0.025 |
| 174 |
257101_at |
AT3G25020
|
AtRLP42, receptor like protein 42, RBPG1, response to Botrytis polygalacturonases 1, RLP42, receptor like protein 42 |
0.233 |
-0.310 |
| 175 |
267483_at |
AT2G02810
|
ATUTR1, UDP-GALACTOSE TRANSPORTER 1, UTR1, UDP-galactose transporter 1 |
0.233 |
-0.181 |
| 176 |
249581_at |
AT5G37600
|
ATGLN1;1, ARABIDOPSIS GLUTAMINE SYNTHASE 1;1, ATGSR1, ARABIDOPSIS THALIANA GLUTAMINE SYNTHASE CLONE R1, GLN1;1, GLUTAMINE SYNTHASE 1;1, GSR 1, glutamine synthase clone R1 |
0.231 |
-0.337 |
| 177 |
263082_at |
AT2G27200
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.231 |
-0.143 |
| 178 |
264767_at |
AT1G61380
|
SD1-29, S-domain-1 29 |
0.231 |
-0.396 |
| 179 |
264211_at |
AT1G22770
|
FB, GI, GIGANTEA |
0.230 |
-0.361 |
| 180 |
266831_at |
AT2G22830
|
SQE2, squalene epoxidase 2 |
0.229 |
-0.364 |
| 181 |
249719_at |
AT5G35735
|
[Auxin-responsive family protein] |
0.229 |
-0.050 |
| 182 |
265510_at |
AT2G05630
|
ATG8D |
0.228 |
-0.186 |
| 183 |
262504_at |
AT1G21750
|
ATPDI5, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 5, ATPDIL1-1, PDI-like 1-1, PDI5, PROTEIN DISULFIDE ISOMERASE 5, PDIL1-1, PDI-like 1-1 |
0.228 |
-0.157 |
| 184 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
0.228 |
-0.699 |
| 185 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
0.227 |
0.096 |
| 186 |
263249_at |
AT2G31360
|
ADS2, 16:0delta9 desaturase 2, AtADS2 |
0.227 |
-0.283 |
| 187 |
261039_at |
AT1G17455
|
ELF4-L4, ELF4-like 4 |
0.225 |
-0.524 |
| 188 |
252362_at |
AT3G48500
|
PDE312, PIGMENT DEFECTIVE 312, PTAC10, PLASTID TRANSCRIPTIONALLY ACTIVE 10, TAC10 |
0.224 |
-0.237 |
| 189 |
266873_at |
AT2G44740
|
CYCP4;1, cyclin p4;1 |
0.224 |
-0.242 |
| 190 |
248510_at |
AT5G50315
|
[Mutator-like transposase family, has a 5.0e-14 P-value blast match to Q9XE24 /118-277 Pfam PF03108 MuDR family transposase (MuDr-element domain)] |
0.222 |
-0.104 |
| 191 |
256930_at |
AT3G22460
|
OASA2, O-acetylserine (thiol) lyase (OAS-TL) isoform A2 |
0.222 |
0.024 |
| 192 |
254248_at |
AT4G23270
|
CRK19, cysteine-rich RLK (RECEPTOR-like protein kinase) 19 |
0.221 |
-0.080 |
| 193 |
265450_at |
AT2G46620
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.221 |
-0.168 |
| 194 |
263864_at |
AT2G04530
|
CPZ, TRZ2, TRNASE Z 2 |
0.221 |
-0.230 |
| 195 |
246782_at |
AT5G27320
|
ATGID1C, GID1C, GA INSENSITIVE DWARF1C |
0.221 |
-0.030 |
| 196 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
0.221 |
-0.088 |
| 197 |
246055_at |
AT5G08380
|
AGAL1, alpha-galactosidase 1, AtAGAL1, alpha-galactosidase 1 |
0.220 |
-0.180 |
| 198 |
254208_at |
AT4G24175
|
unknown |
0.220 |
-0.370 |
| 199 |
249422_at |
AT5G39760
|
AtHB23, homeobox protein 23, HB23, homeobox protein 23, ZHD10, ZINC FINGER HOMEODOMAIN 10 |
0.220 |
-0.341 |
| 200 |
259364_at |
AT1G13260
|
EDF4, ETHYLENE RESPONSE DNA BINDING FACTOR 4, RAV1, related to ABI3/VP1 1 |
0.220 |
-0.110 |
| 201 |
267412_at |
AT2G34940
|
BP80-3;2, binding protein of 80 kDa 3;2, VSR3;2, VACUOLAR SORTING RECEPTOR 3;2, VSR5, VACUOLAR SORTING RECEPTOR 5 |
0.219 |
-0.069 |
| 202 |
245569_at |
AT4G14660
|
NRPE7 |
0.218 |
-0.346 |
| 203 |
260170_at |
AT1G71890
|
ATSUC5, SUCROSE-PROTON SYMPORTER 5, SUC5 |
0.218 |
0.250 |
| 204 |
247043_at |
AT5G66880
|
SNRK2-3, SUCROSE NONFERMENTING 1 (SNF1)-RELATED PROTEIN KINASE 2-3, SNRK2.3, sucrose nonfermenting 1(SNF1)-related protein kinase 2.3, SRK2I |
0.218 |
-0.387 |
| 205 |
266225_at |
AT2G28900
|
ATOEP16-1, outer plastid envelope protein 16-1, ATOEP16-L, OUTER PLASTID ENVELOPE PROTEIN 16-L, OEP16, outer envelope protein 16, OEP16-1, outer plastid envelope protein 16-1 |
0.217 |
-0.542 |
| 206 |
249362_at |
AT5G40550
|
AtSGF29b, SGF29b, SaGa associated Factor 29 b |
0.216 |
-0.132 |
| 207 |
254032_at |
AT4G25940
|
[ENTH/ANTH/VHS superfamily protein] |
0.216 |
-0.179 |
| 208 |
249580_at |
AT5G37740
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.215 |
-0.031 |
| 209 |
253622_at |
AT4G30560
|
ATCNGC9, cyclic nucleotide gated channel 9, CNGC9, cyclic nucleotide gated channel 9 |
0.215 |
-0.211 |
| 210 |
266229_at |
AT2G28840
|
XBAT31, XB3 ortholog 1 in Arabidopsis thaliana |
0.214 |
-0.445 |
| 211 |
248870_at |
AT5G46710
|
[PLATZ transcription factor family protein] |
0.213 |
-0.411 |
| 212 |
246018_at |
AT5G10695
|
unknown |
0.213 |
-0.106 |
| 213 |
249015_at |
AT5G44730
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.213 |
-0.132 |
| 214 |
259303_at |
AT3G05130
|
unknown |
0.213 |
-0.236 |
| 215 |
260799_at |
AT1G78270
|
AtUGT85A4, UDP-glucosyl transferase 85A4, UGT85A4, UDP-glucosyl transferase 85A4 |
0.212 |
-0.084 |
| 216 |
245602_at |
AT4G14270
|
unknown |
0.212 |
-0.651 |
| 217 |
264599_at |
AT1G04600
|
ATXIA, XIA, myosin XI A |
0.212 |
-0.027 |
| 218 |
250194_at |
AT5G14550
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
0.212 |
-0.489 |
| 219 |
259911_at |
AT1G72680
|
ATCAD1, CINNAMYL ALCOHOL DEHYDROGENASE 1, CAD1, cinnamyl-alcohol dehydrogenase |
0.212 |
0.262 |
| 220 |
253987_at |
AT4G26270
|
PFK3, phosphofructokinase 3 |
0.210 |
0.037 |
| 221 |
262607_at |
AT1G13990
|
unknown |
0.210 |
-0.020 |
| 222 |
262296_at |
AT1G27630
|
CYCT1;3, cyclin T 1;3 |
0.209 |
-0.320 |
| 223 |
249974_at |
AT5G18780
|
[F-box/RNI-like superfamily protein] |
0.207 |
-0.064 |
| 224 |
266596_at |
AT2G46150
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.207 |
0.039 |
| 225 |
256597_at |
AT3G28500
|
[60S acidic ribosomal protein family] |
0.206 |
-0.092 |
| 226 |
249220_at |
AT5G42420
|
[Nucleotide-sugar transporter family protein] |
0.205 |
-0.359 |
| 227 |
259534_at |
AT1G12290
|
[Disease resistance protein (CC-NBS-LRR class) family] |
0.205 |
0.083 |
| 228 |
252357_at |
AT3G48410
|
[alpha/beta-Hydrolases superfamily protein] |
0.204 |
-0.068 |
| 229 |
267246_at |
AT2G30250
|
ATWRKY25, WRKY25, WRKY DNA-binding protein 25 |
0.204 |
-0.056 |
| 230 |
253575_at |
AT4G31060
|
[Integrase-type DNA-binding superfamily protein] |
0.204 |
-0.148 |
| 231 |
246791_at |
AT5G27280
|
[Zim17-type zinc finger protein] |
0.203 |
-0.191 |
| 232 |
253636_at |
AT4G30500
|
unknown |
0.203 |
-0.268 |
| 233 |
267378_at |
AT2G26200
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.203 |
-0.352 |
| 234 |
250595_at |
AT5G07770
|
AtFH16, FH16, formin homolog 16 |
0.203 |
-0.086 |
| 235 |
254253_at |
AT4G23320
|
CRK24, cysteine-rich RLK (RECEPTOR-like protein kinase) 24 |
0.202 |
0.155 |
| 236 |
246755_at |
AT5G27920
|
[F-box family protein] |
0.202 |
-0.200 |
| 237 |
246155_at |
AT5G20030
|
[Plant Tudor-like RNA-binding protein] |
0.202 |
-0.148 |
| 238 |
262920_at |
AT1G79500
|
AtkdsA1 |
0.201 |
-0.288 |
| 239 |
245600_at |
AT4G14230
|
unknown |
0.201 |
-0.390 |
| 240 |
255477_at |
AT4G02370
|
unknown |
0.201 |
-0.294 |
| 241 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
0.201 |
-0.401 |
| 242 |
256100_at |
AT1G13750
|
[Purple acid phosphatases superfamily protein] |
0.200 |
-0.217 |
| 243 |
264071_at |
AT2G27920
|
SCPL51, serine carboxypeptidase-like 51 |
0.199 |
-0.493 |
| 244 |
257712_at |
AT3G27420
|
unknown |
0.199 |
0.050 |
| 245 |
257830_at |
AT3G26690
|
ATNUDT13, ARABIDOPSIS THALIANA NUDIX HYDROLASE HOMOLOG 13, ATNUDX13, nudix hydrolase homolog 13, NUDX13, nudix hydrolase homolog 13 |
0.199 |
0.073 |
| 246 |
256837_at |
AT3G22900
|
NRPD7 |
0.199 |
-0.304 |
| 247 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
0.198 |
-0.167 |
| 248 |
247776_at |
AT5G58700
|
ATPLC4, PLC4, phosphatidylinositol-speciwc phospholipase C4 |
0.198 |
0.038 |
| 249 |
250775_at |
AT5G05460
|
AtENGase85A, ENGase85A, Endo-beta-N-acetyglucosaminidase 85A |
0.197 |
-0.078 |
| 250 |
247302_at |
AT5G63880
|
VPS20.1 |
0.197 |
-0.251 |
| 251 |
253253_at |
AT4G34750
|
SAUR49, SMALL AUXIN UPREGULATED RNA 49 |
0.197 |
-0.312 |
| 252 |
260211_at |
AT1G74440
|
unknown |
0.197 |
-0.153 |
| 253 |
264257_at |
AT1G09230
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.197 |
0.277 |
| 254 |
247248_at |
AT5G64560
|
ATMGT9, MGT9, magnesium transporter 9, MRS2-2 |
0.195 |
-0.084 |
| 255 |
251730_at |
AT3G56330
|
[N2,N2-dimethylguanosine tRNA methyltransferase] |
0.195 |
-0.343 |
| 256 |
248276_at |
AT5G53550
|
ATYSL3, YELLOW STRIPE LIKE 3, YSL3, YELLOW STRIPE like 3 |
0.195 |
-0.028 |
| 257 |
265987_at |
AT2G24240
|
[BTB/POZ domain with WD40/YVTN repeat-like protein] |
0.195 |
-0.244 |
| 258 |
245937_at |
AT5G19750
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
0.194 |
-0.108 |
| 259 |
251235_at |
AT3G62860
|
[alpha/beta-Hydrolases superfamily protein] |
0.194 |
0.045 |
| 260 |
252117_at |
AT3G51430
|
SSL5, STRICTOSIDINE SYNTHASE-LIKE 5, YLS2, YELLOW-LEAF-SPECIFIC GENE 2 |
0.194 |
0.078 |
| 261 |
251422_at |
AT3G60540
|
[Preprotein translocase Sec, Sec61-beta subunit protein] |
0.193 |
-0.075 |
| 262 |
264957_at |
AT1G77000
|
ATSKP2;2, ARABIDOPSIS HOMOLOG OF HOMOLOG OF HUMAN SKP2 2, SKP2B |
0.193 |
-0.125 |
| 263 |
259118_at |
AT3G01310
|
[Phosphoglycerate mutase-like family protein] |
0.192 |
-0.424 |
| 264 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
0.192 |
-0.226 |
| 265 |
252281_at |
AT3G49320
|
[Metal-dependent protein hydrolase] |
0.192 |
-0.183 |
| 266 |
255980_at |
AT1G33970
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.192 |
-0.421 |
| 267 |
260671_at |
AT1G19310
|
[RING/U-box superfamily protein] |
0.192 |
-0.134 |
| 268 |
256256_at |
AT3G11230
|
[Yippee family putative zinc-binding protein] |
0.192 |
-0.078 |
| 269 |
263183_at |
AT1G05570
|
ATGSL06, ATGSL6, CALS1, callose synthase 1, GSL06, GSL6, GLUCAN SYNTHASE-LIKE 6 |
0.191 |
-0.105 |
| 270 |
256858_at |
AT3G15140
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
0.191 |
-0.099 |
| 271 |
253553_at |
AT4G31050
|
LIP2, octanoyl- transferase |
0.190 |
-0.398 |
| 272 |
254632_at |
AT4G18630
|
unknown |
0.190 |
-0.077 |
| 273 |
260169_at |
AT1G71990
|
ATFT4, ARABIDOPSIS FUCOSYLTRANSFERASE 4, ATFUT13, FT4-M, FUCTC, FUT13, fucosyltransferase 13 |
0.189 |
-0.129 |
| 274 |
252043_at |
AT3G52390
|
[TatD related DNase] |
0.189 |
-0.141 |
| 275 |
247966_at |
AT5G56610
|
[Phosphotyrosine protein phosphatases superfamily protein] |
0.188 |
-0.190 |
| 276 |
255181_at |
AT4G08110
|
[CACTA-like transposase family (Ptta/En/Spm), has a 4.2e-66 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-0.684 |
0.042 |
| 277 |
257272_at |
AT3G28130
|
UMAMIT44, Usually multiple acids move in and out Transporters 44 |
-0.674 |
0.113 |
| 278 |
256603_at |
AT3G28270
|
unknown |
-0.634 |
1.086 |
| 279 |
259432_at |
AT1G01520
|
ASG4, ALTERED SEED GERMINATION 4 |
-0.611 |
0.721 |
| 280 |
257714_at |
AT3G27360
|
H3.1, histone 3.1 |
-0.575 |
0.070 |
| 281 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
-0.558 |
0.513 |
| 282 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-0.554 |
1.282 |
| 283 |
255694_at |
AT4G00050
|
UNE10, unfertilized embryo sac 10 |
-0.552 |
0.592 |
| 284 |
261684_at |
AT1G47400
|
unknown |
-0.531 |
-0.190 |
| 285 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
-0.528 |
0.490 |
| 286 |
255484_at |
AT4G02540
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.523 |
-0.122 |
| 287 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
-0.516 |
0.570 |
| 288 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
-0.512 |
1.056 |
| 289 |
256303_at |
AT1G69550
|
[disease resistance protein (TIR-NBS-LRR class)] |
-0.497 |
-0.094 |
| 290 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
-0.462 |
0.837 |
| 291 |
254024_at |
AT4G25780
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.437 |
0.168 |
| 292 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
-0.433 |
-0.348 |
| 293 |
247055_at |
AT5G66740
|
unknown |
-0.428 |
0.672 |
| 294 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
-0.427 |
0.251 |
| 295 |
265111_at |
AT1G62510
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.426 |
0.142 |
| 296 |
266841_at |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
-0.426 |
0.022 |
| 297 |
258228_at |
AT3G27610
|
[Nucleotidylyl transferase superfamily protein] |
-0.422 |
0.050 |
| 298 |
257151_at |
AT3G27200
|
[Cupredoxin superfamily protein] |
-0.421 |
-0.014 |
| 299 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
-0.413 |
0.319 |
| 300 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
-0.412 |
1.015 |
| 301 |
259373_at |
AT1G69160
|
unknown |
-0.409 |
0.265 |
| 302 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
-0.402 |
0.671 |
| 303 |
258139_at |
AT3G24520
|
AT-HSFC1, HSFC1, heat shock transcription factor C1 |
-0.397 |
0.345 |
| 304 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
-0.395 |
1.099 |
| 305 |
248549_at |
AT5G50310
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.391 |
-0.007 |
| 306 |
256304_at |
AT1G69523
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.390 |
-0.133 |
| 307 |
254201_at |
AT4G24130
|
unknown |
-0.389 |
0.335 |
| 308 |
266503_at |
AT2G47780
|
[Rubber elongation factor protein (REF)] |
-0.378 |
0.064 |
| 309 |
255891_at |
AT1G17870
|
ATEGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3, EGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3 |
-0.372 |
0.185 |
| 310 |
253074_at |
AT4G36140
|
[disease resistance protein (TIR-NBS-LRR class), putative] |
-0.372 |
-0.044 |
| 311 |
264341_at |
AT1G70270
|
unknown |
-0.372 |
0.077 |
| 312 |
254996_at |
AT4G10390
|
[Protein kinase superfamily protein] |
-0.369 |
0.417 |
| 313 |
249645_at |
AT5G36910
|
THI2.2, thionin 2.2 |
-0.365 |
0.199 |
| 314 |
245692_at |
AT5G04150
|
BHLH101 |
-0.365 |
-0.449 |
| 315 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
-0.359 |
0.018 |
| 316 |
259953_at |
AT1G74810
|
BOR5, REQUIRES HIGH BORON 5 |
-0.358 |
0.077 |
| 317 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
-0.357 |
0.165 |
| 318 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
-0.357 |
0.575 |
| 319 |
257285_at |
AT3G29760
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.355 |
0.009 |
| 320 |
259985_at |
AT1G76620
|
unknown |
-0.353 |
0.014 |
| 321 |
246966_at |
AT5G24850
|
CRY3, cryptochrome 3 |
-0.352 |
0.613 |
| 322 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
-0.351 |
0.779 |
| 323 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
-0.351 |
1.273 |
| 324 |
254773_at |
AT4G13410
|
ATCSLA15, CSLA15, CELLULOSE SYNTHASE LIKE A15 |
-0.351 |
0.793 |
| 325 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
-0.347 |
0.718 |
| 326 |
247463_at |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
-0.346 |
-0.032 |
| 327 |
247461_at |
AT5G62100
|
ATBAG2, BCL-2-associated athanogene 2, BAG2, BCL-2-associated athanogene 2 |
-0.345 |
0.168 |
| 328 |
252412_at |
AT3G47295
|
unknown |
-0.342 |
0.314 |
| 329 |
245816_at |
AT1G26210
|
ATSOFL1, SOB FIVE-LIKE 1, SOFL1, SOB five-like 1 |
-0.341 |
0.023 |
| 330 |
266996_at |
AT2G34490
|
CYP710A2, cytochrome P450, family 710, subfamily A, polypeptide 2 |
-0.340 |
0.132 |
| 331 |
257741_at |
AT3G27340
|
unknown |
-0.338 |
0.083 |
| 332 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
-0.335 |
0.946 |
| 333 |
245449_at |
AT4G16870
|
[copia-like retrotransposon family, has a 0. P-value blast match to dbj|BAA78426.1| polyprotein (AtRE2-1) (Arabidopsis thaliana) (Ty1_Copia-element)] |
-0.331 |
-0.321 |
| 334 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
-0.329 |
0.053 |
| 335 |
249583_at |
AT5G37770
|
CML24, CALMODULIN-LIKE 24, TCH2, TOUCH 2 |
-0.328 |
0.230 |
| 336 |
259209_at |
AT3G09160
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.328 |
-0.083 |
| 337 |
246597_at |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
-0.322 |
0.543 |
| 338 |
257642_at |
AT3G25710
|
ATAIG1, BHLH32, basic helix-loop-helix 32, TMO5, TARGET OF MONOPTEROS 5 |
-0.322 |
0.096 |
| 339 |
254770_at |
AT4G13340
|
LRX3, leucine-rich repeat/extensin 3 |
-0.322 |
0.070 |
| 340 |
255723_at |
AT3G29575
|
AFP3, ABI five binding protein 3 |
-0.321 |
0.190 |
| 341 |
264318_at |
AT1G04220
|
KCS2, 3-ketoacyl-CoA synthase 2 |
-0.319 |
0.038 |
| 342 |
256021_at |
AT1G58270
|
ZW9 |
-0.317 |
0.125 |
| 343 |
263433_at |
AT2G22240
|
ATIPS2, INOSITOL 3-PHOSPHATE SYNTHASE 2, ATMIPS2, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 2, MIPS2, myo-inositol-1-phosphate synthase 2 |
-0.316 |
0.610 |
| 344 |
253571_at |
AT4G31000
|
[Calmodulin-binding protein] |
-0.315 |
0.106 |
| 345 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
-0.315 |
0.920 |
| 346 |
262883_at |
AT1G64780
|
AMT1;2, ammonium transporter 1;2, ATAMT1;2, ammonium transporter 1;2 |
-0.312 |
0.898 |
| 347 |
266424_at |
AT2G41330
|
[Glutaredoxin family protein] |
-0.312 |
0.140 |
| 348 |
262947_at |
AT1G75750
|
GASA1, GAST1 protein homolog 1 |
-0.310 |
0.119 |
| 349 |
249011_at |
AT5G44670
|
GALS2, galactan synthase 2 |
-0.309 |
0.472 |
| 350 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
-0.309 |
0.082 |
| 351 |
264001_at |
AT2G22420
|
[Peroxidase superfamily protein] |
-0.309 |
0.203 |
| 352 |
256582_at |
AT3G28840
|
unknown |
-0.309 |
0.111 |
| 353 |
261907_at |
AT1G65060
|
4CL3, 4-coumarate:CoA ligase 3 |
-0.307 |
0.358 |
| 354 |
256302_at |
AT1G69526
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.306 |
-0.016 |
| 355 |
247109_at |
AT5G65870
|
ATPSK5, phytosulfokine 5 precursor, PSK5, PSK5, phytosulfokine 5 precursor |
-0.306 |
0.334 |
| 356 |
246540_at |
AT5G15600
|
SP1L4, SPIRAL1-like4 |
-0.304 |
0.409 |
| 357 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
-0.300 |
1.084 |
| 358 |
251165_at |
AT3G63340
|
[Protein phosphatase 2C family protein] |
-0.299 |
0.108 |
| 359 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.299 |
0.804 |
| 360 |
245227_s_at |
AT1G08410
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.298 |
-0.107 |
| 361 |
254586_at |
AT4G19510
|
[Disease resistance protein (TIR-NBS-LRR class)] |
-0.297 |
0.118 |
| 362 |
257051_at |
AT3G15270
|
SPL5, squamosa promoter binding protein-like 5 |
-0.294 |
0.422 |
| 363 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
-0.292 |
0.934 |
| 364 |
256980_at |
AT3G26932
|
DRB3, dsRNA-binding protein 3 |
-0.292 |
0.054 |
| 365 |
250828_at |
AT5G05250
|
unknown |
-0.290 |
-0.184 |
| 366 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
-0.289 |
0.051 |
| 367 |
263545_at |
AT2G21560
|
unknown |
-0.288 |
0.280 |
| 368 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.286 |
0.417 |
| 369 |
256674_at |
AT3G52360
|
unknown |
-0.285 |
0.152 |
| 370 |
260955_at |
AT1G06000
|
[UDP-Glycosyltransferase superfamily protein] |
-0.284 |
0.307 |
| 371 |
253332_at |
AT4G33420
|
[Peroxidase superfamily protein] |
-0.283 |
0.182 |
| 372 |
265122_at |
AT1G62540
|
FMO GS-OX2, flavin-monooxygenase glucosinolate S-oxygenase 2 |
-0.282 |
0.184 |
| 373 |
259103_at |
AT3G11690
|
unknown |
-0.282 |
0.243 |
| 374 |
252160_at |
AT3G50570
|
[hydroxyproline-rich glycoprotein family protein] |
-0.281 |
0.173 |
| 375 |
251859_at |
AT3G54680
|
[proteophosphoglycan-related] |
-0.281 |
0.210 |
| 376 |
256275_at |
AT3G12110
|
ACT11, actin-11 |
-0.280 |
0.373 |
| 377 |
253165_at |
AT4G35320
|
unknown |
-0.279 |
0.214 |
| 378 |
261150_at |
AT1G19640
|
JMT, jasmonic acid carboxyl methyltransferase |
-0.279 |
0.855 |
| 379 |
258315_at |
AT3G16175
|
[Thioesterase superfamily protein] |
-0.278 |
0.437 |
| 380 |
259149_at |
AT3G10340
|
PAL4, phenylalanine ammonia-lyase 4 |
-0.278 |
0.234 |
| 381 |
250528_at |
AT5G08600
|
[U3 ribonucleoprotein (Utp) family protein] |
-0.277 |
-0.115 |
| 382 |
264102_at |
AT1G79270
|
ECT8, evolutionarily conserved C-terminal region 8 |
-0.277 |
0.799 |
| 383 |
254683_at |
AT4G13800
|
unknown |
-0.276 |
0.276 |
| 384 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
-0.275 |
0.802 |
| 385 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
-0.273 |
0.926 |
| 386 |
255028_at |
AT4G09890
|
unknown |
-0.269 |
0.295 |
| 387 |
264052_at |
AT2G22330
|
CYP79B3, cytochrome P450, family 79, subfamily B, polypeptide 3 |
-0.267 |
0.197 |
| 388 |
249588_at |
AT5G37790
|
[Protein kinase superfamily protein] |
-0.266 |
0.128 |
| 389 |
267497_at |
AT2G30540
|
[Thioredoxin superfamily protein] |
-0.265 |
0.429 |
| 390 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
-0.263 |
1.117 |
| 391 |
263918_at |
AT2G36590
|
ATPROT3, PROLINE TRANSPORTER 3, ProT3, proline transporter 3 |
-0.262 |
0.269 |
| 392 |
247030_at |
AT5G67210
|
IRX15-L, IRX15-LIKE |
-0.261 |
0.573 |
| 393 |
265573_at |
AT2G28200
|
[C2H2-type zinc finger family protein] |
-0.259 |
0.611 |
| 394 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.258 |
0.806 |
| 395 |
257801_at |
AT3G18750
|
ATWNK6, ARABIDOPSIS THALIANA WITH NO K 6, WNK6, with no lysine (K) kinase 6, ZIK5 |
-0.258 |
0.475 |
| 396 |
247038_at |
AT5G67160
|
EPS1, ENHANCED PSEUDOMONAS SUSCEPTIBILTY 1 |
-0.257 |
0.467 |
| 397 |
252937_at |
AT4G39180
|
ATSEC14, ARABIDOPSIS THALIANA SECRETION 14, SEC14, SECRETION 14 |
-0.256 |
-0.085 |
| 398 |
245558_at |
AT4G15430
|
[ERD (early-responsive to dehydration stress) family protein] |
-0.254 |
0.610 |
| 399 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.252 |
0.489 |
| 400 |
247175_at |
AT5G65280
|
GCL1, GCR2-like 1 |
-0.251 |
0.425 |
| 401 |
264692_at |
AT1G70000
|
[myb-like transcription factor family protein] |
-0.250 |
0.465 |
| 402 |
251966_at |
AT3G53380
|
LecRK-VIII.1, L-type lectin receptor kinase VIII.1 |
-0.249 |
0.077 |
| 403 |
253814_at |
AT4G28290
|
unknown |
-0.249 |
0.369 |
| 404 |
256527_at |
AT1G66100
|
[Plant thionin] |
-0.249 |
0.310 |
| 405 |
263320_at |
AT2G47180
|
AtGolS1, galactinol synthase 1, GolS1, galactinol synthase 1 |
-0.249 |
1.059 |
| 406 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
-0.248 |
0.735 |
| 407 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.246 |
0.635 |
| 408 |
258736_at |
AT3G05900
|
[neurofilament protein-related] |
-0.245 |
0.242 |
| 409 |
265892_at |
AT2G15020
|
unknown |
-0.243 |
0.882 |
| 410 |
259169_at |
AT3G03520
|
NPC3, non-specific phospholipase C3 |
-0.243 |
0.201 |
| 411 |
245262_at |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
-0.243 |
-0.035 |
| 412 |
255563_at |
AT4G01740
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.242 |
-0.083 |
| 413 |
262098_at |
AT1G56170
|
ATHAP5B, HAP5B, NF-YC2, nuclear factor Y, subunit C2 |
-0.241 |
0.474 |
| 414 |
253859_at |
AT4G27657
|
unknown |
-0.240 |
0.440 |
| 415 |
259856_at |
AT1G68440
|
unknown |
-0.239 |
0.602 |
| 416 |
263486_at |
AT2G22200
|
[Integrase-type DNA-binding superfamily protein] |
-0.239 |
0.195 |
| 417 |
245479_at |
AT4G16140
|
[proline-rich family protein] |
-0.239 |
0.542 |
| 418 |
251050_at |
AT5G02440
|
unknown |
-0.239 |
0.163 |
| 419 |
263796_at |
AT2G24540
|
AFR, ATTENUATED FAR-RED RESPONSE |
-0.238 |
0.625 |
| 420 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
-0.238 |
0.722 |
| 421 |
263467_at |
AT2G31730
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.238 |
-0.047 |
| 422 |
252427_at |
AT3G47640
|
PYE, POPEYE |
-0.237 |
0.084 |
| 423 |
246142_at |
AT5G19970
|
unknown |
-0.237 |
0.258 |
| 424 |
255064_at |
AT4G08950
|
EXO, EXORDIUM |
-0.236 |
0.160 |
| 425 |
266661_at |
AT2G25820
|
ESE2, ethylene and salt inducible 2 |
-0.236 |
0.345 |
| 426 |
246075_at |
AT5G20410
|
ATMGD2, ARABIDOPSIS THALIANA MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE 2, MGD2, monogalactosyldiacylglycerol synthase 2 |
-0.236 |
0.137 |
| 427 |
254575_at |
AT4G19460
|
[UDP-Glycosyltransferase superfamily protein] |
-0.236 |
0.259 |
| 428 |
256725_at |
AT2G34070
|
TBL37, TRICHOME BIREFRINGENCE-LIKE 37 |
-0.235 |
0.456 |
| 429 |
260603_at |
AT1G55960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.235 |
0.650 |
| 430 |
252097_at |
AT3G51090
|
unknown |
-0.235 |
0.254 |
| 431 |
251827_at |
AT3G55120
|
A11, AtCHI, CFI, CHALCONE FLAVANONE ISOMERASE, CHI, chalcone isomerase, TT5, TRANSPARENT TESTA 5 |
-0.234 |
0.165 |
| 432 |
260337_at |
AT1G69310
|
ATWRKY57, WRKY57, WRKY DNA-binding protein 57 |
-0.233 |
0.117 |
| 433 |
262212_at |
AT1G74890
|
ARR15, response regulator 15 |
-0.232 |
0.239 |
| 434 |
257013_at |
AT3G26922
|
[F-box/RNI-like superfamily protein] |
-0.232 |
-0.048 |
| 435 |
264909_at |
AT2G17300
|
unknown |
-0.231 |
0.166 |
| 436 |
253263_at |
AT4G34000
|
ABF3, abscisic acid responsive elements-binding factor 3, AtABF3, DPBF5, DC3 PROMOTER-BINDING FACTOR 5 |
-0.231 |
0.398 |
| 437 |
246516_at |
AT5G15740
|
[O-fucosyltransferase family protein] |
-0.230 |
0.289 |
| 438 |
245386_at |
AT4G14010
|
RALFL32, ralf-like 32 |
-0.230 |
0.107 |
| 439 |
250558_at |
AT5G07990
|
CYP75B1, CYTOCHROME P450 75B1, D501, TT7, TRANSPARENT TESTA 7 |
-0.229 |
0.062 |
| 440 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
-0.227 |
0.686 |
| 441 |
249798_at |
AT5G23730
|
EFO2, EARLY FLOWERING BY OVEREXPRESSION 2, RUP2, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 2 |
-0.226 |
0.451 |
| 442 |
249063_at |
AT5G44110
|
ABCI21, ATP-binding cassette A21, ATNAP2, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN, ATPOP1, POP1 |
-0.225 |
0.379 |
| 443 |
252917_at |
AT4G38960
|
BBX19, B-box domain protein 19 |
-0.224 |
0.539 |
| 444 |
249910_at |
AT5G22630
|
ADT5, arogenate dehydratase 5 |
-0.223 |
0.200 |
| 445 |
250141_at |
AT5G14640
|
ATSK13, SHAGGY-LIKE KINASE 13, SK13, shaggy-like kinase 13 |
-0.223 |
0.204 |
| 446 |
258021_at |
AT3G19380
|
PUB25, plant U-box 25 |
-0.222 |
0.324 |
| 447 |
251420_at |
AT3G60490
|
[Integrase-type DNA-binding superfamily protein] |
-0.222 |
0.183 |
| 448 |
249797_at |
AT5G23750
|
[Remorin family protein] |
-0.221 |
0.121 |
| 449 |
260297_at |
AT1G80280
|
[alpha/beta-Hydrolases superfamily protein] |
-0.220 |
-0.097 |
| 450 |
254041_at |
AT4G25830
|
[Uncharacterised protein family (UPF0497)] |
-0.220 |
0.190 |
| 451 |
260840_at |
AT1G29050
|
TBL38, TRICHOME BIREFRINGENCE-LIKE 38 |
-0.218 |
0.305 |
| 452 |
247779_at |
AT5G58760
|
DDB2, damaged DNA binding 2 |
-0.217 |
0.377 |
| 453 |
267166_at |
AT2G37720
|
TBL15, TRICHOME BIREFRINGENCE-LIKE 15 |
-0.217 |
-0.032 |
| 454 |
253579_at |
AT4G30610
|
BRS1, BRI1 SUPPRESSOR 1, SCPL24, SERINE CARBOXYPEPTIDASE 24 PRECURSOR |
-0.216 |
0.535 |
| 455 |
261832_at |
AT1G10650
|
[SBP (S-ribonuclease binding protein) family protein] |
-0.216 |
0.259 |
| 456 |
257270_at |
AT3G28140
|
[RNA ligase/cyclic nucleotide phosphodiesterase family protein] |
-0.215 |
0.231 |
| 457 |
254508_at |
AT4G20170
|
GALS3, galactan synthase 3 |
-0.214 |
0.150 |
| 458 |
259934_at |
AT1G71340
|
AtGDPD4, GDPD4, glycerophosphodiester phosphodiesterase 4 |
-0.213 |
0.136 |
| 459 |
255957_at |
AT1G22160
|
unknown |
-0.213 |
0.040 |
| 460 |
266391_at |
AT2G41290
|
SSL2, strictosidine synthase-like 2 |
-0.212 |
0.545 |
| 461 |
248091_at |
AT5G55120
|
galactose-1-phosphate guanylyltransferase (GDP)s;GDP-D-glucose phosphorylases;quercetin 4'-O-glucosyltransferases |
-0.212 |
0.072 |
| 462 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
-0.211 |
0.499 |
| 463 |
259834_at |
AT1G69570
|
[Dof-type zinc finger DNA-binding family protein] |
-0.211 |
0.498 |
| 464 |
263632_at |
AT2G04795
|
unknown |
-0.211 |
0.271 |
| 465 |
249765_at |
AT5G24030
|
SLAH3, SLAC1 homologue 3 |
-0.209 |
0.163 |
| 466 |
262376_at |
AT1G72970
|
EDA17, embryo sac development arrest 17, HTH, HOTHEAD |
-0.209 |
0.082 |
| 467 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
-0.209 |
0.486 |
| 468 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
-0.209 |
0.312 |
| 469 |
264466_at |
AT1G10380
|
[Putative membrane lipoprotein] |
-0.209 |
-0.055 |
| 470 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
-0.209 |
0.155 |
| 471 |
251727_at |
AT3G56290
|
unknown |
-0.208 |
0.621 |
| 472 |
256093_at |
AT1G20823
|
[RING/U-box superfamily protein] |
-0.208 |
0.246 |
| 473 |
250955_at |
AT5G03190
|
CPUORF47, conserved peptide upstream open reading frame 47 |
-0.208 |
0.268 |
| 474 |
255549_at |
AT4G01950
|
ATGPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3, GPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3 |
-0.207 |
0.135 |
| 475 |
245346_at |
AT4G17090
|
AtBAM3, BAM3, BETA-AMYLASE 3, BMY8, BETA-AMYLASE 8, CT-BMY, chloroplast beta-amylase |
-0.207 |
0.201 |
| 476 |
248404_at |
AT5G51460
|
ATTPPA, TPPA, trehalose-6-phosphate phosphatase A |
-0.205 |
0.238 |
| 477 |
251975_at |
AT3G53230
|
AtCDC48B, cell division cycle 48B |
-0.205 |
0.306 |
| 478 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
-0.204 |
0.212 |
| 479 |
254817_at |
AT4G12460
|
ORP2B, OSBP(oxysterol binding protein)-related protein 2B |
-0.204 |
-0.036 |
| 480 |
253264_at |
AT4G33950
|
ATOST1, OPEN STOMATA 1, OST1, OPEN STOMATA 1, P44, SNRK2-6, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-6, SNRK2.6, SNF1-RELATED PROTEIN KINASE 2.6, SRK2E |
-0.204 |
0.219 |
| 481 |
254496_at |
AT4G20070
|
AAH, allantoate amidohydrolase, ATAAH, allantoate amidohydrolase |
-0.203 |
0.258 |
| 482 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
-0.203 |
0.592 |
| 483 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
-0.203 |
0.396 |
| 484 |
250248_at |
AT5G13740
|
ZIF1, zinc induced facilitator 1 |
-0.202 |
0.026 |
| 485 |
254332_at |
AT4G22730
|
[Leucine-rich repeat protein kinase family protein] |
-0.202 |
0.059 |
| 486 |
245141_at |
AT2G45400
|
BEN1, BRI1-5 ENHANCED 1 |
-0.201 |
0.075 |
| 487 |
262912_at |
AT1G59740
|
AtNPF4.3, NPF4.3, NRT1/ PTR family 4.3 |
-0.201 |
0.129 |
| 488 |
249622_at |
AT5G37550
|
unknown |
-0.200 |
0.235 |
| 489 |
255818_at |
AT2G33570
|
GALS1, galactan synthase 1 |
-0.200 |
0.216 |
| 490 |
256764_at |
AT3G29310
|
[calmodulin-binding protein-related] |
-0.200 |
-0.118 |
| 491 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
-0.199 |
-0.084 |
| 492 |
255576_at |
AT4G01440
|
UMAMIT31, Usually multiple acids move in and out Transporters 31 |
-0.198 |
0.182 |
| 493 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
-0.198 |
0.523 |
| 494 |
259922_at |
AT1G72770
|
AtHAB1, HAB1, HYPERSENSITIVE TO ABA1 |
-0.198 |
0.391 |
| 495 |
266418_at |
AT2G38750
|
ANNAT4, annexin 4, AtANN4 |
-0.197 |
0.426 |
| 496 |
246247_at |
AT4G36640
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.197 |
0.213 |
| 497 |
253638_at |
AT4G30470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.197 |
0.154 |
| 498 |
261109_at |
AT1G75450
|
ATCKX5, ARABIDOPSIS THALIANA CYTOKININ OXIDASE 5, ATCKX6, CYTOKININ OXIDASE 6, CKX5, cytokinin oxidase 5 |
-0.196 |
0.438 |
| 499 |
246495_at |
AT5G16200
|
[50S ribosomal protein-related] |
-0.196 |
0.480 |
| 500 |
263845_at |
AT2G37040
|
ATPAL1, PAL1, PHE ammonia lyase 1 |
-0.194 |
0.102 |
| 501 |
261350_at |
AT1G79770
|
unknown |
-0.193 |
0.439 |
| 502 |
264436_at |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
-0.193 |
0.735 |
| 503 |
245777_at |
AT1G73540
|
atnudt21, nudix hydrolase homolog 21, NUDT21, nudix hydrolase homolog 21 |
-0.193 |
-0.003 |
| 504 |
252363_at |
AT3G48460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.193 |
0.486 |
| 505 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
-0.193 |
0.614 |
| 506 |
248080_at |
AT5G55380
|
[MBOAT (membrane bound O-acyl transferase) family protein] |
-0.192 |
0.327 |
| 507 |
245676_at |
AT1G56670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.192 |
0.188 |
| 508 |
251370_at |
AT3G60450
|
[Phosphoglycerate mutase family protein] |
-0.191 |
-0.043 |
| 509 |
248395_at |
AT5G52120
|
AtPP2-A14, phloem protein 2-A14, PP2-A14, phloem protein 2-A14 |
-0.191 |
0.262 |
| 510 |
263192_at |
AT1G36160
|
ACC1, acetyl-CoA carboxylase 1, AT-ACC1, EMB22, EMBRYO DEFECTIVE 22, GK, GURKE, GSD1, GLOSSYHEAD 1, PAS3, PASTICCINO 3, SFR3, SENSITIVE TO FREEZING 3 |
-0.191 |
0.303 |
| 511 |
264822_at |
AT1G03457
|
AtBRN2, BRN2, Bruno-like 2 |
-0.191 |
0.090 |
| 512 |
263134_at |
AT1G78570
|
ATRHM1, ARABIDOPSIS THALIANA RHAMNOSE BIOSYNTHESIS 1, RHM1, rhamnose biosynthesis 1, ROL1, REPRESSOR OF LRX1 1 |
-0.191 |
0.063 |
| 513 |
264862_at |
AT1G24330
|
[ARM repeat superfamily protein] |
-0.191 |
0.207 |
| 514 |
248205_at |
AT5G54300
|
unknown |
-0.190 |
0.446 |
| 515 |
263629_at |
AT2G04850
|
[Auxin-responsive family protein] |
-0.190 |
0.105 |
| 516 |
257216_at |
AT3G14990
|
AtDJ1A, DJ-1 homolog A, DJ-1a, DJ1A, DJ-1 homolog A |
-0.190 |
0.197 |
| 517 |
266447_at |
AT2G43290
|
MSS3, multicopy suppressors of snf4 deficiency in yeast 3 |
-0.189 |
0.349 |
| 518 |
249278_at |
AT5G41900
|
[alpha/beta-Hydrolases superfamily protein] |
-0.189 |
0.071 |
| 519 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
-0.189 |
0.241 |
| 520 |
248270_at |
AT5G53450
|
ORG1, OBP3-responsive gene 1 |
-0.189 |
-0.309 |
| 521 |
262737_at |
AT1G28560
|
SRD2, SHOOT REDIFFERENTIATION DEFECTIVE 2 |
-0.188 |
0.125 |
| 522 |
254145_at |
AT4G24700
|
unknown |
-0.187 |
0.619 |
| 523 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-0.187 |
0.397 |
| 524 |
245427_at |
AT4G17550
|
AtG3Pp4, glycerol-3-phosphate permease 4, G3Pp4, glycerol-3-phosphate permease 4 |
-0.186 |
0.269 |
| 525 |
249415_at |
AT5G39660
|
CDF2, cycling DOF factor 2 |
-0.186 |
0.368 |
| 526 |
261926_at |
AT1G22530
|
PATL2, PATELLIN 2 |
-0.186 |
0.010 |
| 527 |
251476_at |
AT3G59670
|
unknown |
-0.185 |
0.159 |
| 528 |
256564_at |
AT3G29770
|
ATMES11, ARABIDOPSIS THALIANA METHYL ESTERASE 11, MES11, methyl esterase 11 |
-0.185 |
0.297 |
| 529 |
250335_at |
AT5G11650
|
[alpha/beta-Hydrolases superfamily protein] |
-0.185 |
0.258 |
| 530 |
251494_at |
AT3G59350
|
[Protein kinase superfamily protein] |
-0.185 |
-0.347 |
| 531 |
255732_at |
AT1G25450
|
CER60, ECERIFERUM 60, KCS5, 3-ketoacyl-CoA synthase 5 |
-0.185 |
-0.251 |
| 532 |
252419_at |
AT3G47510
|
unknown |
-0.185 |
-0.053 |
| 533 |
261924_at |
AT1G22550
|
[Major facilitator superfamily protein] |
-0.184 |
-0.083 |
| 534 |
245296_at |
AT4G16370
|
oligopeptide transporter |
-0.184 |
0.023 |
| 535 |
251544_at |
AT3G58790
|
GAUT15, galacturonosyltransferase 15 |
-0.184 |
0.093 |
| 536 |
244974_at |
ATCG00700
|
PSBN, photosystem II reaction center protein N |
-0.182 |
-0.023 |
| 537 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
-0.182 |
0.280 |
| 538 |
246494_at |
AT5G16190
|
ATCSLA11, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE A11, CSLA11, cellulose synthase like A11 |
-0.181 |
0.262 |
| 539 |
265634_at |
AT2G25530
|
[AFG1-like ATPase family protein] |
-0.181 |
0.306 |
| 540 |
261958_at |
AT1G64500
|
[Glutaredoxin family protein] |
-0.181 |
1.068 |
| 541 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.180 |
0.294 |
| 542 |
264040_at |
AT2G03730
|
ACR5, ACT domain repeat 5 |
-0.180 |
0.342 |
| 543 |
246069_at |
AT5G20220
|
[zinc knuckle (CCHC-type) family protein] |
-0.180 |
0.469 |
| 544 |
254020_at |
AT4G25700
|
B1, BCH1, BETA CAROTENOID HYDROXYLASE 1, BETA-OHASE 1, beta-hydroxylase 1, chy1 |
-0.180 |
0.126 |
| 545 |
253824_at |
AT4G27940
|
ATMTM1, ARABIDOPSIS MANGANESE TRACKING FACTOR FOR MITOCHONDRIAL SOD2, MTM1, manganese tracking factor for mitochondrial SOD2 |
-0.179 |
0.180 |
| 546 |
267066_at |
AT2G41040
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.179 |
0.353 |
| 547 |
248236_at |
AT5G53870
|
AtENODL1, ENODL1, early nodulin-like protein 1 |
-0.179 |
-0.087 |
| 548 |
259794_at |
AT1G64330
|
[myosin heavy chain-related] |
-0.178 |
-0.063 |
| 549 |
254065_at |
AT4G25420
|
AT2301, ATGA20OX1, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 1, GA20OX1, GA5, GA REQUIRING 5 |
-0.178 |
0.212 |