|
probeID |
AGICode |
Annotation |
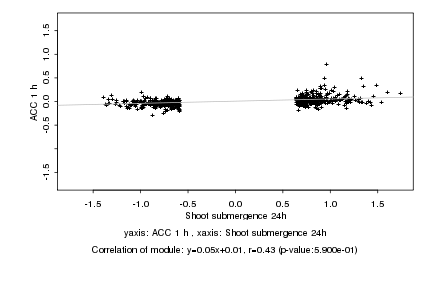
Log2 signal ratio
Shoot submergence 24h |
Log2 signal ratio
ACC 1 h |
| 1 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
1.736 |
0.185 |
| 2 |
260668_at |
AT1G19530
|
unknown |
1.599 |
0.205 |
| 3 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
1.530 |
-0.014 |
| 4 |
260527_at |
AT2G47270
|
UPB1, UPBEAT1 |
1.484 |
0.341 |
| 5 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
1.451 |
0.108 |
| 6 |
250666_at |
AT5G07100
|
WRKY26, WRKY DNA-binding protein 26 |
1.431 |
-0.071 |
| 7 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
1.418 |
-0.007 |
| 8 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
1.401 |
0.006 |
| 9 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
1.377 |
-0.026 |
| 10 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
1.344 |
0.329 |
| 11 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
1.329 |
-0.011 |
| 12 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
1.324 |
0.488 |
| 13 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
1.319 |
-0.014 |
| 14 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
1.311 |
0.068 |
| 15 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
1.299 |
-0.000 |
| 16 |
258383_at |
AT3G15440
|
unknown |
1.297 |
0.021 |
| 17 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
1.296 |
0.053 |
| 18 |
251012_at |
AT5G02580
|
unknown |
1.265 |
0.030 |
| 19 |
249454_at |
AT5G39520
|
unknown |
1.253 |
0.120 |
| 20 |
256743_at |
AT3G29370
|
P1R3, P1R3 |
1.216 |
0.057 |
| 21 |
254001_at |
AT4G26260
|
MIOX4, myo-inositol oxygenase 4 |
1.213 |
-0.003 |
| 22 |
250182_at |
AT5G14470
|
AtGALK2, GALK2, galactokinase 2 |
1.207 |
0.028 |
| 23 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
1.198 |
0.044 |
| 24 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
1.197 |
0.062 |
| 25 |
246925_at |
AT5G25180
|
CYP71B14, cytochrome P450, family 71, subfamily B, polypeptide 14 |
1.186 |
0.021 |
| 26 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
1.185 |
0.034 |
| 27 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
1.184 |
0.073 |
| 28 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
1.184 |
-0.015 |
| 29 |
247954_at |
AT5G56870
|
BGAL4, beta-galactosidase 4 |
1.179 |
-0.003 |
| 30 |
260741_at |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
1.179 |
0.153 |
| 31 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
1.177 |
0.213 |
| 32 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
1.170 |
-0.021 |
| 33 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
1.169 |
-0.130 |
| 34 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
1.162 |
0.145 |
| 35 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
1.162 |
0.020 |
| 36 |
245233_at |
AT4G25580
|
[CAP160 protein] |
1.157 |
0.019 |
| 37 |
253829_at |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
1.150 |
0.117 |
| 38 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
1.146 |
-0.082 |
| 39 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
1.142 |
0.095 |
| 40 |
252320_at |
AT3G48580
|
XTH11, xyloglucan endotransglucosylase/hydrolase 11 |
1.139 |
0.083 |
| 41 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
1.137 |
0.085 |
| 42 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
1.135 |
0.064 |
| 43 |
247919_at |
AT5G57650
|
[eukaryotic translation initiation factor-related] |
1.122 |
0.044 |
| 44 |
246072_at |
AT5G20240
|
PI, PISTILLATA |
1.109 |
0.061 |
| 45 |
260662_at |
AT1G19540
|
[NmrA-like negative transcriptional regulator family protein] |
1.098 |
0.025 |
| 46 |
253373_at |
AT4G33150
|
LKR, LKR/SDH, LYSINE-KETOGLUTARATE REDUCTASE/SACCHAROPINE DEHYDROGENASE, SDH, SACCHAROPINE DEHYDROGENASE |
1.097 |
0.034 |
| 47 |
266392_at |
AT2G41280
|
ATM10, M10 |
1.096 |
0.040 |
| 48 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
1.091 |
0.164 |
| 49 |
258005_at |
AT3G19390
|
[Granulin repeat cysteine protease family protein] |
1.088 |
0.054 |
| 50 |
257670_at |
AT3G20340
|
unknown |
1.080 |
-0.042 |
| 51 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
1.072 |
0.135 |
| 52 |
264580_at |
AT1G05340
|
unknown |
1.063 |
0.048 |
| 53 |
252539_at |
AT3G45730
|
unknown |
1.061 |
0.057 |
| 54 |
250940_at |
AT5G03310
|
SAUR44, SMALL AUXIN UPREGULATED RNA 44 |
1.058 |
0.045 |
| 55 |
252993_at |
AT4G38540
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
1.053 |
0.041 |
| 56 |
245317_at |
AT4G15610
|
[Uncharacterised protein family (UPF0497)] |
1.049 |
0.001 |
| 57 |
262643_at |
AT1G62770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
1.047 |
0.094 |
| 58 |
258225_at |
AT3G15630
|
unknown |
1.047 |
0.042 |
| 59 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
1.045 |
0.017 |
| 60 |
247878_at |
AT5G57760
|
unknown |
1.034 |
0.297 |
| 61 |
252511_at |
AT3G46280
|
[protein kinase-related] |
1.033 |
0.070 |
| 62 |
263475_at |
AT2G31945
|
unknown |
1.032 |
0.217 |
| 63 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
1.026 |
0.033 |
| 64 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
1.022 |
0.013 |
| 65 |
267036_at |
AT2G38465
|
unknown |
1.013 |
-0.040 |
| 66 |
265539_at |
AT2G15830
|
unknown |
1.012 |
0.239 |
| 67 |
264467_at |
AT1G10140
|
[Uncharacterised conserved protein UCP031279] |
1.012 |
0.070 |
| 68 |
258395_at |
AT3G15500
|
ANAC055, NAC domain containing protein 55, ATNAC3, NAC domain containing protein 3, NAC055, NAC domain containing protein 55, NAC3, NAC domain containing protein 3 |
1.011 |
0.022 |
| 69 |
259841_at |
AT1G52200
|
[PLAC8 family protein] |
0.993 |
0.023 |
| 70 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.991 |
-0.002 |
| 71 |
250680_at |
AT5G06570
|
[alpha/beta-Hydrolases superfamily protein] |
0.986 |
0.190 |
| 72 |
260748_at |
AT1G49210
|
[RING/U-box superfamily protein] |
0.983 |
0.188 |
| 73 |
246238_at |
AT4G36670
|
AtPLT6, AtPMT6, PLT6, polyol transporter 6, PMT6, polyol/monosaccharide transporter 6 |
0.969 |
0.008 |
| 74 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
0.967 |
0.157 |
| 75 |
249869_at |
AT5G23050
|
AAE17, acyl-activating enzyme 17 |
0.964 |
0.002 |
| 76 |
248895_at |
AT5G46330
|
FLS2, FLAGELLIN-SENSITIVE 2 |
0.961 |
0.180 |
| 77 |
265266_at |
AT2G42890
|
AML2, MEI2-like 2, ML2, MEI2-like 2 |
0.960 |
-0.039 |
| 78 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
0.958 |
0.786 |
| 79 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
0.957 |
0.077 |
| 80 |
264124_at |
AT1G79360
|
ATOCT2, organic cation/carnitine transporter 2, OCT2, ORGANIC CATION TRANSPORTER 2, OCT2, organic cation/carnitine transporter 2 |
0.955 |
0.069 |
| 81 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
0.954 |
0.031 |
| 82 |
261567_at |
AT1G33055
|
unknown |
0.952 |
0.137 |
| 83 |
246251_at |
AT4G37220
|
[Cold acclimation protein WCOR413 family] |
0.951 |
0.067 |
| 84 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.951 |
0.103 |
| 85 |
259982_at |
AT1G76410
|
ATL8 |
0.942 |
0.269 |
| 86 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
0.942 |
-0.022 |
| 87 |
264616_at |
AT2G17740
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.938 |
0.018 |
| 88 |
262325_at |
AT1G64160
|
AtDIR5, DIR5, dirigent protein 5 |
0.938 |
0.066 |
| 89 |
249364_at |
AT5G40590
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.938 |
0.494 |
| 90 |
251647_at |
AT3G57770
|
[Protein kinase superfamily protein] |
0.937 |
0.002 |
| 91 |
248208_at |
AT5G53980
|
ATHB52, homeobox protein 52, HB52, homeobox protein 52 |
0.931 |
0.353 |
| 92 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
0.920 |
0.024 |
| 93 |
263194_at |
AT1G36060
|
[Integrase-type DNA-binding superfamily protein] |
0.920 |
0.016 |
| 94 |
263281_at |
AT2G14160
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.918 |
0.018 |
| 95 |
252081_at |
AT3G51910
|
AT-HSFA7A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A7A, HSFA7A, heat shock transcription factor A7A |
0.917 |
0.057 |
| 96 |
249783_at |
AT5G24270
|
ATSOS3, SALT OVERLY SENSITIVE 3, CBL4, CALCINEURIN B-LIKE PROTEIN 4, SOS3, SALT OVERLY SENSITIVE 3 |
0.915 |
-0.004 |
| 97 |
259058_at |
AT3G03470
|
CYP89A9, cytochrome P450, family 87, subfamily A, polypeptide 9 |
0.912 |
0.045 |
| 98 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
0.906 |
-0.118 |
| 99 |
247774_at |
AT5G58660
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.905 |
0.114 |
| 100 |
245696_at |
AT5G04190
|
PKS4, phytochrome kinase substrate 4 |
0.902 |
0.290 |
| 101 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
0.902 |
-0.016 |
| 102 |
251026_at |
AT5G02200
|
FHL, far-red-elongated hypocotyl1-like |
0.901 |
0.064 |
| 103 |
248509_at |
AT5G50335
|
unknown |
0.895 |
0.282 |
| 104 |
264238_at |
AT1G54740
|
unknown |
0.895 |
0.006 |
| 105 |
263653_at |
AT1G04310
|
ERS2, ethylene response sensor 2 |
0.893 |
0.289 |
| 106 |
266259_at |
AT2G27830
|
unknown |
0.892 |
0.048 |
| 107 |
257922_at |
AT3G23150
|
ETR2, ethylene response 2 |
0.891 |
0.178 |
| 108 |
266477_at |
AT2G31085
|
AtCLE6, CLE6, CLAVATA3/ESR-RELATED 6 |
0.889 |
0.033 |
| 109 |
255381_at |
AT4G03510
|
ATRMA1, RMA1, RING membrane-anchor 1 |
0.888 |
0.112 |
| 110 |
267230_at |
AT2G44080
|
ARL, ARGOS-like |
0.887 |
0.327 |
| 111 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
0.887 |
-0.037 |
| 112 |
250951_at |
AT5G03550
|
unknown |
0.886 |
0.047 |
| 113 |
259076_at |
AT3G02140
|
AFP4, ABI FIVE BINDING PROTEIN 4, TMAC2, TWO OR MORE ABRES-CONTAINING GENE 2 |
0.882 |
-0.045 |
| 114 |
249955_at |
AT5G18840
|
[Major facilitator superfamily protein] |
0.880 |
0.116 |
| 115 |
264513_at |
AT1G09420
|
G6PD4, glucose-6-phosphate dehydrogenase 4 |
0.878 |
-0.063 |
| 116 |
247800_at |
AT5G58570
|
unknown |
0.874 |
0.059 |
| 117 |
247136_at |
AT5G66170
|
STR18, sulfurtransferase 18 |
0.874 |
0.008 |
| 118 |
255905_at |
AT1G17810
|
BETA-TIP, beta-tonoplast intrinsic protein |
0.872 |
0.008 |
| 119 |
253322_at |
AT4G33980
|
unknown |
0.871 |
0.043 |
| 120 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
0.870 |
-0.046 |
| 121 |
252570_at |
AT3G45300
|
ATIVD, IVD, isovaleryl-CoA-dehydrogenase, IVDH, ISOVALERYL-COA-DEHYDROGENASE |
0.869 |
-0.151 |
| 122 |
263169_at |
AT1G03010
|
[Phototropic-responsive NPH3 family protein] |
0.868 |
0.164 |
| 123 |
251116_at |
AT3G63470
|
scpl40, serine carboxypeptidase-like 40 |
0.866 |
-0.042 |
| 124 |
252136_at |
AT3G50770
|
CML41, calmodulin-like 41 |
0.863 |
0.033 |
| 125 |
259120_at |
AT3G02240
|
GLV4, GOLVEN 4, RGF7, root meristem growth factor 7 |
0.862 |
-0.015 |
| 126 |
263118_at |
AT1G03090
|
MCCA |
0.860 |
-0.026 |
| 127 |
256697_at |
AT3G20660
|
AtOCT4, organic cation/carnitine transporter4, OCT4, organic cation/carnitine transporter4 |
0.856 |
-0.081 |
| 128 |
265481_at |
AT2G15960
|
unknown |
0.855 |
0.078 |
| 129 |
251356_at |
AT3G61060
|
AtPP2-A13, phloem protein 2-A13, PP2-A13, phloem protein 2-A13 |
0.854 |
-0.131 |
| 130 |
248564_at |
AT5G49700
|
AHL17, AT-hook motif nuclear localized protein 17 |
0.854 |
0.233 |
| 131 |
261272_at |
AT1G26665
|
[Mediator complex, subunit Med10] |
0.852 |
0.010 |
| 132 |
260926_at |
AT1G21360
|
GLTP2, glycolipid transfer protein 2 |
0.851 |
0.178 |
| 133 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
0.849 |
-0.023 |
| 134 |
260451_at |
AT1G72360
|
AtERF73, ERF73, ethylene response factor 73, HRE1, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 1 |
0.846 |
0.111 |
| 135 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.845 |
0.108 |
| 136 |
264482_at |
AT1G77210
|
AtSTP14, sugar transport protein 14, STP14, sugar transport protein 14 |
0.845 |
-0.021 |
| 137 |
256061_at |
AT1G07040
|
unknown |
0.845 |
0.032 |
| 138 |
252924_at |
AT4G39070
|
BBX20, B-box domain protein 20, BZS1, BZS1 |
0.844 |
0.022 |
| 139 |
256965_at |
AT3G13450
|
DIN4, DARK INDUCIBLE 4 |
0.840 |
-0.060 |
| 140 |
250152_at |
AT5G15120
|
unknown |
0.839 |
0.052 |
| 141 |
254500_at |
AT4G20110
|
BP80-3;1, binding protein of 80 kDa 3;1, VSR3;1, VACUOLAR SORTING RECEPTOR 3;1, VSR7, VACUOLAR SORTING RECEPTOR 7 |
0.838 |
-0.044 |
| 142 |
260101_at |
AT1G73260
|
ATKTI1, ARABIDOPSIS THALIANA KUNITZ TRYPSIN INHIBITOR 1, KTI1, kunitz trypsin inhibitor 1 |
0.836 |
0.021 |
| 143 |
261586_at |
AT1G01640
|
[BTB/POZ domain-containing protein] |
0.836 |
-0.111 |
| 144 |
258227_at |
AT3G15620
|
UVR3, UV REPAIR DEFECTIVE 3 |
0.833 |
0.001 |
| 145 |
267178_at |
AT2G37750
|
unknown |
0.830 |
0.063 |
| 146 |
246114_at |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
0.829 |
0.036 |
| 147 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.824 |
0.069 |
| 148 |
247394_at |
AT5G62865
|
unknown |
0.822 |
-0.030 |
| 149 |
254314_at |
AT4G22470
|
[protease inhibitor/seed storage/lipid transfer protein (LTP) family protein] |
0.819 |
-0.011 |
| 150 |
248703_at |
AT5G48430
|
[Eukaryotic aspartyl protease family protein] |
0.819 |
0.128 |
| 151 |
248432_at |
AT5G51390
|
unknown |
0.817 |
0.029 |
| 152 |
255742_at |
AT1G25560
|
AtTEM1, EDF1, ETHYLENE RESPONSE DNA BINDING FACTOR 1, TEM1, TEMPRANILLO 1 |
0.817 |
0.244 |
| 153 |
253827_at |
AT4G28085
|
unknown |
0.814 |
0.018 |
| 154 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
0.811 |
-0.014 |
| 155 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
0.811 |
0.038 |
| 156 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
0.809 |
-0.000 |
| 157 |
247573_at |
AT5G61160
|
AACT1, anthocyanin 5-aromatic acyltransferase 1 |
0.808 |
0.006 |
| 158 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
0.808 |
0.079 |
| 159 |
246929_at |
AT5G25210
|
unknown |
0.806 |
0.093 |
| 160 |
258110_at |
AT3G14610
|
CYP72A7, cytochrome P450, family 72, subfamily A, polypeptide 7 |
0.806 |
-0.036 |
| 161 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.805 |
0.056 |
| 162 |
248193_at |
AT5G54080
|
AtHGO, HGO, homogentisate 1,2-dioxygenase |
0.805 |
-0.023 |
| 163 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
0.803 |
0.179 |
| 164 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
0.803 |
0.016 |
| 165 |
260933_at |
AT1G02470
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.800 |
0.062 |
| 166 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
0.800 |
-0.043 |
| 167 |
261658_at |
AT1G50040
|
unknown |
0.798 |
0.047 |
| 168 |
247474_at |
AT5G62280
|
unknown |
0.798 |
0.217 |
| 169 |
252109_at |
AT3G51540
|
unknown |
0.796 |
0.042 |
| 170 |
266156_at |
AT2G28110
|
FRA8, FRAGILE FIBER 8, IRX7, IRREGULAR XYLEM 7 |
0.796 |
-0.024 |
| 171 |
267385_at |
AT2G44380
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.794 |
0.016 |
| 172 |
246935_at |
AT5G25350
|
EBF2, EIN3-binding F box protein 2 |
0.794 |
0.196 |
| 173 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
0.794 |
0.010 |
| 174 |
267075_at |
AT2G41070
|
ATBZIP12, DPBF4, EEL, ENHANCED EM LEVEL |
0.793 |
0.017 |
| 175 |
261211_at |
AT1G12780
|
ATUGE1, A. THALIANA UDP-GLC 4-EPIMERASE 1, UGE1, UDP-D-glucose/UDP-D-galactose 4-epimerase 1 |
0.790 |
-0.013 |
| 176 |
247128_at |
AT5G66110
|
HIPP27, heavy metal associated isoprenylated plant protein 27 |
0.789 |
0.072 |
| 177 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
0.787 |
0.041 |
| 178 |
258327_at |
AT3G22640
|
PAP85 |
0.786 |
-0.021 |
| 179 |
258362_at |
AT3G14280
|
unknown |
0.785 |
0.016 |
| 180 |
255543_at |
AT4G01870
|
[tolB protein-related] |
0.784 |
0.088 |
| 181 |
264434_at |
AT1G10340
|
[Ankyrin repeat family protein] |
0.783 |
0.044 |
| 182 |
249415_at |
AT5G39660
|
CDF2, cycling DOF factor 2 |
0.781 |
-0.046 |
| 183 |
252068_at |
AT3G51440
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.781 |
-0.031 |
| 184 |
259244_at |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
0.781 |
0.002 |
| 185 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
0.780 |
0.003 |
| 186 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
0.777 |
0.005 |
| 187 |
246984_at |
AT5G67310
|
CYP81G1, cytochrome P450, family 81, subfamily G, polypeptide 1 |
0.771 |
0.099 |
| 188 |
245393_at |
AT4G16260
|
[Glycosyl hydrolase superfamily protein] |
0.769 |
0.039 |
| 189 |
245228_at |
AT3G29810
|
COBL2, COBRA-like protein 2 precursor |
0.768 |
-0.106 |
| 190 |
258059_at |
AT3G29035
|
ANAC059, Arabidopsis NAC domain containing protein 59, ATNAC3, NAC domain containing protein 3, NAC3, NAC domain containing protein 3, ORS1, ORE1 SISTER1 |
0.766 |
0.156 |
| 191 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
0.764 |
0.051 |
| 192 |
262805_at |
AT1G20900
|
AHL27, AT-hook motif nuclear-localized protein 27, ESC, ESCAROLA, ORE7, ORESARA 7 |
0.759 |
-0.020 |
| 193 |
256097_at |
AT1G13670
|
unknown |
0.758 |
0.015 |
| 194 |
246831_at |
AT5G26340
|
ATSTP13, SUGAR TRANSPORT PROTEIN 13, MSS1, STP13, SUGAR TRANSPORT PROTEIN 13 |
0.757 |
-0.068 |
| 195 |
254409_at |
AT4G21400
|
CRK28, cysteine-rich RLK (RECEPTOR-like protein kinase) 28 |
0.755 |
0.073 |
| 196 |
267628_at |
AT2G42280
|
AKS3, ABA-responsive kinase substrate 3, FBH4, FLOWERING BHLH 4 |
0.753 |
0.165 |
| 197 |
252183_at |
AT3G50740
|
UGT72E1, UDP-glucosyl transferase 72E1 |
0.749 |
0.025 |
| 198 |
247951_at |
AT5G57240
|
ORP4C, OSBP(oxysterol binding protein)-related protein 4C |
0.747 |
0.063 |
| 199 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
0.746 |
-0.085 |
| 200 |
251621_at |
AT3G57700
|
[Protein kinase superfamily protein] |
0.744 |
0.079 |
| 201 |
262607_at |
AT1G13990
|
unknown |
0.743 |
-0.070 |
| 202 |
248040_at |
AT5G55970
|
[RING/U-box superfamily protein] |
0.742 |
0.071 |
| 203 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.741 |
0.005 |
| 204 |
265536_at |
AT2G15880
|
[Leucine-rich repeat (LRR) family protein] |
0.740 |
0.200 |
| 205 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
0.740 |
0.027 |
| 206 |
266983_at |
AT2G39400
|
[alpha/beta-Hydrolases superfamily protein] |
0.739 |
0.239 |
| 207 |
266839_at |
AT2G25930
|
ELF3, EARLY FLOWERING 3, PYK20 |
0.739 |
-0.014 |
| 208 |
255283_at |
AT4G04620
|
ATG8B, autophagy 8b |
0.737 |
-0.054 |
| 209 |
250649_at |
AT5G06690
|
WCRKC1, WCRKC thioredoxin 1 |
0.737 |
0.017 |
| 210 |
264887_at |
AT1G23120
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.735 |
-0.080 |
| 211 |
265511_at |
AT2G05540
|
[Glycine-rich protein family] |
0.734 |
0.108 |
| 212 |
264902_at |
AT1G23060
|
MDP40, MICROTUBULE DESTABILIZING PROTEIN 40 |
0.734 |
0.086 |
| 213 |
248911_at |
AT5G45830
|
ATDOG1, DOG1, DELAY OF GERMINATION 1, GAAS5, germination ability after storage 5, GSQ5, GLUCOSE SENSING QTL 5 |
0.730 |
0.108 |
| 214 |
251479_at |
AT3G59700
|
ATHLECRK, lectin-receptor kinase, HLECRK, lectin-receptor kinase, LecRK-V.5, L-type lectin receptor kinase V.5, LecRK-V.5, LEGUME-LIKE LECTIN RECEPTOR KINASE-V.5, LECRK1, LECTIN-RECEPTOR KINASE 1 |
0.730 |
-0.119 |
| 215 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.729 |
0.147 |
| 216 |
266140_at |
AT2G28120
|
[Major facilitator superfamily protein] |
0.728 |
-0.019 |
| 217 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
0.727 |
-0.032 |
| 218 |
246028_at |
AT5G21170
|
5'-AMP-activated protein kinase beta-2 subunit protein |
0.722 |
-0.009 |
| 219 |
250777_at |
AT5G05440
|
PYL5, PYRABACTIN RESISTANCE 1-LIKE 5, RCAR8, regulatory component of ABA receptor 8 |
0.721 |
0.089 |
| 220 |
264209_at |
AT1G22740
|
ATRABG3B, RAB7, RAB75, RABG3B, RAB GTPase homolog G3B |
0.721 |
-0.090 |
| 221 |
260581_at |
AT2G47190
|
ATMYB2, MYB DOMAIN PROTEIN 2, MYB2, myb domain protein 2 |
0.717 |
-0.014 |
| 222 |
264246_at |
AT1G60140
|
ATTPS10, trehalose phosphate synthase, TPS10, trehalose phosphate synthase, TPS10, TREHALOSE PHOSPHATE SYNTHASE 10 |
0.716 |
-0.056 |
| 223 |
247793_at |
AT5G58650
|
PSY1, plant peptide containing sulfated tyrosine 1 |
0.716 |
0.010 |
| 224 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.715 |
0.016 |
| 225 |
258833_at |
AT3G07274
|
|
0.712 |
0.128 |
| 226 |
254197_at |
AT4G24040
|
ATTRE1, TRE1, trehalase 1 |
0.711 |
0.033 |
| 227 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.710 |
-0.089 |
| 228 |
253281_at |
AT4G34138
|
UGT73B1, UDP-glucosyl transferase 73B1 |
0.709 |
-0.093 |
| 229 |
248969_at |
AT5G45310
|
unknown |
0.706 |
0.036 |
| 230 |
245794_at |
AT1G32170
|
XTH30, xyloglucan endotransglucosylase/hydrolase 30, XTR4, xyloglucan endotransglycosylase 4 |
0.705 |
-0.012 |
| 231 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
0.705 |
0.168 |
| 232 |
262981_at |
AT1G75590
|
SAUR52, SMALL AUXIN UPREGULATED RNA 52 |
0.704 |
0.170 |
| 233 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
0.704 |
0.012 |
| 234 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
0.704 |
0.038 |
| 235 |
254797_at |
AT4G13030
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.703 |
-0.007 |
| 236 |
260011_at |
AT1G68110
|
[ENTH/ANTH/VHS superfamily protein] |
0.702 |
0.032 |
| 237 |
247013_at |
AT5G67480
|
ATBT4, BT4, BTB and TAZ domain protein 4 |
0.700 |
-0.013 |
| 238 |
258939_at |
AT3G10020
|
unknown |
0.700 |
-0.079 |
| 239 |
262322_at |
AT1G27590
|
unknown |
0.699 |
0.049 |
| 240 |
254954_at |
AT4G10910
|
unknown |
0.697 |
0.121 |
| 241 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
0.697 |
-0.041 |
| 242 |
251039_at |
AT5G02020
|
SIS, Salt Induced Serine rich |
0.696 |
0.114 |
| 243 |
252464_at |
AT3G47160
|
[RING/U-box superfamily protein] |
0.696 |
0.073 |
| 244 |
259694_at |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
0.691 |
-0.081 |
| 245 |
246396_at |
AT1G58180
|
ATBCA6, A. THALIANA BETA CARBONIC ANHYDRASE 6, BCA6, beta carbonic anhydrase 6 |
0.690 |
-0.030 |
| 246 |
267532_at |
AT2G42040
|
unknown |
0.690 |
-0.022 |
| 247 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
0.689 |
0.055 |
| 248 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
0.689 |
0.024 |
| 249 |
254726_at |
AT4G13660
|
ATPRR2, PRR2, pinoresinol reductase 2 |
0.689 |
0.066 |
| 250 |
252940_at |
AT4G39270
|
[Leucine-rich repeat protein kinase family protein] |
0.689 |
-0.071 |
| 251 |
265070_at |
AT1G55510
|
BCDH BETA1, branched-chain alpha-keto acid decarboxylase E1 beta subunit |
0.687 |
0.067 |
| 252 |
260639_at |
AT1G53180
|
unknown |
0.685 |
0.094 |
| 253 |
247835_at |
AT5G57910
|
unknown |
0.684 |
0.114 |
| 254 |
253519_at |
AT4G31240
|
AtNRX2, NRX2, nucleoredoxin 2 |
0.684 |
-0.033 |
| 255 |
251245_at |
AT3G62090
|
PIF6, PHYTOCHROME-INTERACTING FACTOR 6, PIL2, phytochrome interacting factor 3-like 2 |
0.681 |
0.038 |
| 256 |
248347_at |
AT5G52250
|
EFO1, EARLY FLOWERING BY OVEREXPRESSION 1, RUP1, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 |
0.680 |
0.013 |
| 257 |
252303_at |
AT3G49210
|
[O-acyltransferase (WSD1-like) family protein] |
0.678 |
-0.080 |
| 258 |
257373_at |
AT2G43140
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.676 |
0.160 |
| 259 |
246951_at |
AT5G04880
|
[expressed protein] |
0.676 |
-0.034 |
| 260 |
247924_at |
AT5G57655
|
[xylose isomerase family protein] |
0.675 |
-0.017 |
| 261 |
254193_at |
AT4G23870
|
unknown |
0.675 |
-0.094 |
| 262 |
265902_at |
AT2G25590
|
[Plant Tudor-like protein] |
0.675 |
0.024 |
| 263 |
249377_at |
AT5G40690
|
unknown |
0.673 |
-0.018 |
| 264 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
0.671 |
0.023 |
| 265 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
0.670 |
0.043 |
| 266 |
249032_at |
AT5G44910
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.669 |
0.049 |
| 267 |
250580_at |
AT5G07440
|
GDH2, glutamate dehydrogenase 2 |
0.666 |
0.025 |
| 268 |
251037_at |
AT5G02100
|
ORP3A, OSBP(OXYSTEROL BINDING PROTEIN)-RELATED PROTEIN 3A, UNE18, UNFERTILIZED EMBRYO SAC 18 |
0.666 |
-0.010 |
| 269 |
264767_at |
AT1G61380
|
SD1-29, S-domain-1 29 |
0.666 |
0.101 |
| 270 |
246195_at |
AT4G36410
|
UBC17, ubiquitin-conjugating enzyme 17 |
0.664 |
0.105 |
| 271 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
0.664 |
-0.042 |
| 272 |
252117_at |
AT3G51430
|
SSL5, STRICTOSIDINE SYNTHASE-LIKE 5, YLS2, YELLOW-LEAF-SPECIFIC GENE 2 |
0.663 |
-0.032 |
| 273 |
252100_at |
AT3G51110
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.663 |
0.061 |
| 274 |
261674_at |
AT1G18270
|
[ketose-bisphosphate aldolase class-II family protein] |
0.663 |
0.002 |
| 275 |
250028_at |
AT5G18130
|
unknown |
0.663 |
-0.046 |
| 276 |
247979_at |
AT5G56750
|
NDL1, N-MYC downregulated-like 1 |
0.661 |
0.040 |
| 277 |
262599_at |
AT1G15350
|
unknown |
0.660 |
0.013 |
| 278 |
262580_at |
AT1G15330
|
[Cystathionine beta-synthase (CBS) protein] |
0.660 |
0.079 |
| 279 |
263363_at |
AT2G03850
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.659 |
0.087 |
| 280 |
265028_at |
AT1G24530
|
[Transducin/WD40 repeat-like superfamily protein] |
0.659 |
0.012 |
| 281 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
0.658 |
-0.170 |
| 282 |
264835_at |
AT1G03550
|
AtSCAMP4, SCAMP4, Secretory carrier membrane protein 4 |
0.656 |
-0.061 |
| 283 |
249754_at |
AT5G24530
|
DMR6, DOWNY MILDEW RESISTANT 6 |
0.656 |
0.026 |
| 284 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.655 |
0.012 |
| 285 |
267384_at |
AT2G44370
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.655 |
0.041 |
| 286 |
251642_at |
AT3G57520
|
AtSIP2, seed imbibition 2, RS2, raffinose synthase 2, SIP2, seed imbibition 2 |
0.652 |
-0.002 |
| 287 |
253401_at |
AT4G32870
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.650 |
-0.009 |
| 288 |
251282_at |
AT3G61630
|
CRF6, cytokinin response factor 6 |
0.650 |
-0.053 |
| 289 |
265471_at |
AT2G37130
|
[Peroxidase superfamily protein] |
0.645 |
-0.010 |
| 290 |
249999_at |
AT5G18640
|
[alpha/beta-Hydrolases superfamily protein] |
0.645 |
0.011 |
| 291 |
251144_at |
AT5G01210
|
[HXXXD-type acyl-transferase family protein] |
0.644 |
0.245 |
| 292 |
255430_at |
AT4G03320
|
AtTic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV, Tic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV |
0.643 |
0.063 |
| 293 |
264045_at |
AT2G22450
|
AtRIBA2, RIBA2, homolog of ribA 2 |
0.643 |
-0.016 |
| 294 |
250213_at |
AT5G13820
|
ATBP-1, ATBP1, ATTBP1, HPPBF-1, H-PROTEIN PROMOTE, TBP1, telomeric DNA binding protein 1 |
0.641 |
0.031 |
| 295 |
249987_at |
AT5G18490
|
unknown |
0.640 |
-0.056 |
| 296 |
253722_at |
AT4G29190
|
AtC3H49, AtOZF2, Arabidopsis thaliana oxidation-related zinc finger 2, AtTZF3, OZF2, oxidation-related zinc finger 2, TZF3, tandem zinc finger 3 |
0.638 |
0.100 |
| 297 |
250208_at |
AT5G14000
|
anac084, NAC domain containing protein 84, NAC084, NAC domain containing protein 84 |
0.638 |
0.068 |
| 298 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
0.637 |
0.053 |
| 299 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.636 |
0.067 |
| 300 |
267436_at |
AT2G19190
|
FRK1, FLG22-induced receptor-like kinase 1 |
0.636 |
0.088 |
| 301 |
256096_at |
AT1G13650
|
unknown |
-1.393 |
0.099 |
| 302 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-1.376 |
-0.058 |
| 303 |
252661_at |
AT3G44450
|
unknown |
-1.362 |
-0.075 |
| 304 |
258055_at |
AT3G16250
|
NDF4, NDH-dependent cyclic electron flow 1, PnsB3, Photosynthetic NDH subcomplex B 3 |
-1.347 |
-0.041 |
| 305 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
-1.347 |
0.047 |
| 306 |
260955_at |
AT1G06000
|
[UDP-Glycosyltransferase superfamily protein] |
-1.338 |
-0.037 |
| 307 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
-1.308 |
0.132 |
| 308 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
-1.305 |
0.050 |
| 309 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
-1.272 |
-0.056 |
| 310 |
260547_at |
AT2G43550
|
[Scorpion toxin-like knottin superfamily protein] |
-1.262 |
-0.016 |
| 311 |
257021_at |
AT3G19710
|
BCAT4, branched-chain aminotransferase4 |
-1.261 |
0.024 |
| 312 |
263829_at |
AT2G40435
|
unknown |
-1.231 |
-0.072 |
| 313 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
-1.216 |
-0.087 |
| 314 |
260745_at |
AT1G78370
|
ATGSTU20, glutathione S-transferase TAU 20, GSTU20, glutathione S-transferase TAU 20 |
-1.190 |
0.009 |
| 315 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
-1.182 |
0.008 |
| 316 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
-1.177 |
0.003 |
| 317 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
-1.173 |
-0.023 |
| 318 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
-1.159 |
0.032 |
| 319 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
-1.153 |
-0.098 |
| 320 |
249774_at |
AT5G24150
|
SQE5, SQUALENE MONOOXYGENASE 5, SQP1 |
-1.150 |
0.008 |
| 321 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
-1.144 |
-0.128 |
| 322 |
252363_at |
AT3G48460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-1.132 |
-0.107 |
| 323 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
-1.125 |
-0.057 |
| 324 |
255786_at |
AT1G19670
|
ATCLH1, chlorophyllase 1, ATHCOR1, CORONATINE-INDUCED PROTEIN 1, CLH1, chlorophyllase 1, CORI1, CORONATINE-INDUCED PROTEIN 1 |
-1.124 |
-0.145 |
| 325 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
-1.107 |
-0.073 |
| 326 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
-1.101 |
-0.072 |
| 327 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
-1.091 |
-0.051 |
| 328 |
254687_at |
AT4G13770
|
CYP83A1, cytochrome P450, family 83, subfamily A, polypeptide 1, REF2, REDUCED EPIDERMAL FLUORESCENCE 2 |
-1.086 |
0.008 |
| 329 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-1.084 |
0.018 |
| 330 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
-1.084 |
0.032 |
| 331 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
-1.082 |
-0.039 |
| 332 |
266326_at |
AT2G46650
|
ATCB5-C, ARABIDOPSIS CYTOCHROME B5 ISOFORM C, B5 #1, CB5-C, cytochrome B5 isoform C, CYTB5-C |
-1.074 |
0.002 |
| 333 |
254371_at |
AT4G21760
|
BGLU47, beta-glucosidase 47 |
-1.068 |
-0.051 |
| 334 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-1.068 |
-0.009 |
| 335 |
264436_at |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
-1.066 |
-0.009 |
| 336 |
250006_at |
AT5G18660
|
PCB2, PALE-GREEN AND CHLOROPHYLL B REDUCED 2 |
-1.062 |
-0.019 |
| 337 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
-1.062 |
-0.090 |
| 338 |
255088_at |
AT4G09350
|
CRRJ, CHLORORESPIRATORY REDUCTION J, DJC75, DNA J protein C75, NdhT, NADH dehydrogenase-like complex T |
-1.050 |
-0.016 |
| 339 |
253971_at |
AT4G26530
|
AtFBA5, FBA5, fructose-bisphosphate aldolase 5 |
-1.044 |
-0.054 |
| 340 |
252073_at |
AT3G51750
|
unknown |
-1.038 |
0.031 |
| 341 |
247463_at |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
-1.037 |
-0.167 |
| 342 |
258675_at |
AT3G08770
|
LTP6, lipid transfer protein 6 |
-1.015 |
-0.010 |
| 343 |
265121_at |
AT1G62560
|
FMO GS-OX3, flavin-monooxygenase glucosinolate S-oxygenase 3 |
-1.007 |
0.007 |
| 344 |
249866_at |
AT5G23010
|
IMS3, 2-ISOPROPYLMALATE SYNTHASE 3, MAM1, methylthioalkylmalate synthase 1 |
-1.003 |
0.017 |
| 345 |
259348_at |
AT3G03770
|
[Leucine-rich repeat protein kinase family protein] |
-0.995 |
-0.123 |
| 346 |
263495_at |
AT2G42530
|
COR15B, cold regulated 15b |
-0.993 |
0.192 |
| 347 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
-0.992 |
-0.143 |
| 348 |
250598_at |
AT5G07690
|
ATMYB29, myb domain protein 29, MYB29, myb domain protein 29, PMG2, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 2 |
-0.991 |
0.012 |
| 349 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
-0.990 |
-0.135 |
| 350 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-0.990 |
-0.098 |
| 351 |
264636_at |
AT1G65490
|
unknown |
-0.987 |
-0.069 |
| 352 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
-0.982 |
-0.041 |
| 353 |
263840_at |
AT2G36885
|
unknown |
-0.981 |
-0.084 |
| 354 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
-0.979 |
-0.117 |
| 355 |
248961_at |
AT5G45650
|
[subtilase family protein] |
-0.976 |
-0.060 |
| 356 |
257793_at |
AT3G26960
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.974 |
0.108 |
| 357 |
261913_at |
AT1G65860
|
FMO GS-OX1, flavin-monooxygenase glucosinolate S-oxygenase 1 |
-0.974 |
-0.037 |
| 358 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
-0.966 |
0.036 |
| 359 |
263287_at |
AT2G36145
|
unknown |
-0.964 |
-0.028 |
| 360 |
248625_at |
AT5G48880
|
KAT5, 3-KETO-ACYL-COENZYME A THIOLASE 5, PKT1, PEROXISOMAL-3-KETO-ACYL-COA THIOLASE 1, PKT2, peroxisomal 3-keto-acyl-CoA thiolase 2 |
-0.962 |
-0.053 |
| 361 |
261150_at |
AT1G19640
|
JMT, jasmonic acid carboxyl methyltransferase |
-0.961 |
0.010 |
| 362 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
-0.959 |
-0.038 |
| 363 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
-0.958 |
-0.055 |
| 364 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
-0.951 |
0.081 |
| 365 |
258396_at |
AT3G15460
|
[Ribosomal RNA processing Brix domain protein] |
-0.939 |
-0.045 |
| 366 |
249614_at |
AT5G37300
|
WSD1 |
-0.937 |
0.105 |
| 367 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
-0.927 |
-0.011 |
| 368 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.926 |
-0.050 |
| 369 |
254862_at |
AT4G12030
|
BASS5, BILE ACID:SODIUM SYMPORTER FAMILY PROTEIN 5, BAT5, bile acid transporter 5 |
-0.926 |
-0.017 |
| 370 |
263841_at |
AT2G36870
|
AtXTH32, XTH32, xyloglucan endotransglucosylase/hydrolase 32 |
-0.925 |
-0.009 |
| 371 |
257057_at |
AT3G15310
|
unknown |
-0.921 |
-0.035 |
| 372 |
262086_at |
AT1G56050
|
[GTP-binding protein-related] |
-0.920 |
-0.014 |
| 373 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
-0.915 |
-0.031 |
| 374 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
-0.912 |
-0.035 |
| 375 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
-0.910 |
-0.055 |
| 376 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-0.900 |
0.023 |
| 377 |
253496_at |
AT4G31870
|
ATGPX7, GLUTATHIONE PEROXIDASE 7, GPX7, glutathione peroxidase 7 |
-0.899 |
-0.034 |
| 378 |
254773_at |
AT4G13410
|
ATCSLA15, CSLA15, CELLULOSE SYNTHASE LIKE A15 |
-0.899 |
0.066 |
| 379 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.898 |
0.069 |
| 380 |
262682_at |
AT1G75900
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.897 |
-0.047 |
| 381 |
256304_at |
AT1G69523
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.897 |
-0.022 |
| 382 |
265892_at |
AT2G15020
|
unknown |
-0.896 |
-0.066 |
| 383 |
251058_at |
AT5G01790
|
unknown |
-0.896 |
-0.072 |
| 384 |
256125_at |
AT1G18250
|
ATLP-1 |
-0.887 |
-0.027 |
| 385 |
264442_at |
AT1G27480
|
[alpha/beta-Hydrolases superfamily protein] |
-0.882 |
-0.042 |
| 386 |
245501_at |
AT4G15620
|
[Uncharacterised protein family (UPF0497)] |
-0.882 |
-0.275 |
| 387 |
252870_at |
AT4G39940
|
AKN2, APS-kinase 2, APK2, ADENOSINE-5'-PHOSPHOSULFATE (APS) KINASE 2 |
-0.880 |
0.039 |
| 388 |
259655_at |
AT1G55210
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.872 |
-0.030 |
| 389 |
261942_at |
AT1G22590
|
AGL87, AGAMOUS-like 87 |
-0.868 |
-0.083 |
| 390 |
264839_at |
AT1G03630
|
POR C, protochlorophyllide oxidoreductase C, PORC |
-0.867 |
0.014 |
| 391 |
255298_at |
AT4G04840
|
ATMSRB6, methionine sulfoxide reductase B6, MSRB6, methionine sulfoxide reductase B6 |
-0.865 |
-0.004 |
| 392 |
262883_at |
AT1G64780
|
AMT1;2, ammonium transporter 1;2, ATAMT1;2, ammonium transporter 1;2 |
-0.865 |
-0.046 |
| 393 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.864 |
-0.126 |
| 394 |
254783_at |
AT4G12830
|
[alpha/beta-Hydrolases superfamily protein] |
-0.863 |
0.030 |
| 395 |
251418_at |
AT3G60440
|
[Phosphoglycerate mutase family protein] |
-0.850 |
-0.021 |
| 396 |
255773_at |
AT1G18590
|
ATSOT17, SULFOTRANSFERASE 17, ATST5C, ARABIDOPSIS SULFOTRANSFERASE 5C, SOT17, sulfotransferase 17 |
-0.848 |
0.031 |
| 397 |
259925_at |
AT1G75040
|
PR-5, PR5, pathogenesis-related gene 5 |
-0.847 |
0.020 |
| 398 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.847 |
-0.092 |
| 399 |
256015_at |
AT1G19150
|
LHCA2*1, Lhca6, photosystem I light harvesting complex gene 6 |
-0.846 |
-0.012 |
| 400 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
-0.846 |
0.066 |
| 401 |
261247_at |
AT1G20070
|
unknown |
-0.845 |
-0.053 |
| 402 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
-0.845 |
-0.118 |
| 403 |
256427_at |
AT3G11090
|
LBD21, LOB domain-containing protein 21 |
-0.844 |
-0.133 |
| 404 |
248867_at |
AT5G46830
|
ATNIG1, NACL-INDUCIBLE GENE 1, NIG1, NACL-inducible gene 1 |
-0.843 |
0.037 |
| 405 |
261330_at |
AT1G44900
|
ATMCM2, MCM2, MINICHROMOSOME MAINTENANCE 2 |
-0.839 |
-0.095 |
| 406 |
263918_at |
AT2G36590
|
ATPROT3, PROLINE TRANSPORTER 3, ProT3, proline transporter 3 |
-0.839 |
0.069 |
| 407 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.833 |
0.031 |
| 408 |
263174_at |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
-0.831 |
-0.067 |
| 409 |
258370_at |
AT3G14395
|
unknown |
-0.831 |
-0.007 |
| 410 |
262109_at |
AT1G02730
|
ATCSLD5, cellulose synthase-like D5, CSLD5, CELLULOSE SYNTHASE LIKE D5, CSLD5, cellulose synthase-like D5, SOS6, SALT OVERLY SENSITIVE 6 |
-0.815 |
-0.018 |
| 411 |
252010_at |
AT3G52740
|
unknown |
-0.811 |
-0.052 |
| 412 |
257533_at |
AT3G10840
|
[alpha/beta-Hydrolases superfamily protein] |
-0.809 |
-0.046 |
| 413 |
247218_at |
AT5G65010
|
ASN2, asparagine synthetase 2 |
-0.806 |
-0.001 |
| 414 |
261016_at |
AT1G26560
|
BGLU40, beta glucosidase 40 |
-0.805 |
-0.049 |
| 415 |
259308_at |
AT3G05180
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.805 |
-0.074 |
| 416 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
-0.800 |
-0.092 |
| 417 |
258859_at |
AT3G02120
|
[hydroxyproline-rich glycoprotein family protein] |
-0.796 |
-0.033 |
| 418 |
248377_at |
AT5G51720
|
At-NEET, NEET, NEET group protein |
-0.794 |
-0.120 |
| 419 |
254233_at |
AT4G23800
|
3xHMG-box2, 3xHigh Mobility Group-box2 |
-0.792 |
-0.039 |
| 420 |
260380_at |
AT1G73870
|
BBX16, B-box domain protein 16, COL7, CONSTANS-LIKE 7 |
-0.784 |
-0.098 |
| 421 |
255016_at |
AT4G10120
|
ATSPS4F |
-0.780 |
-0.025 |
| 422 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
-0.780 |
0.029 |
| 423 |
266395_at |
AT2G43100
|
ATLEUD1, IPMI2, isopropylmalate isomerase 2 |
-0.779 |
0.006 |
| 424 |
262309_at |
AT1G70820
|
[phosphoglucomutase, putative / glucose phosphomutase, putative] |
-0.771 |
-0.011 |
| 425 |
260343_at |
AT1G69200
|
FLN2, fructokinase-like 2 |
-0.769 |
-0.030 |
| 426 |
261921_at |
AT1G65900
|
unknown |
-0.769 |
-0.043 |
| 427 |
261931_at |
AT1G22430
|
[GroES-like zinc-binding dehydrogenase family protein] |
-0.768 |
0.007 |
| 428 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
-0.767 |
-0.033 |
| 429 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.764 |
-0.239 |
| 430 |
247816_at |
AT5G58260
|
NdhN, NADH dehydrogenase-like complex N |
-0.763 |
-0.010 |
| 431 |
253579_at |
AT4G30610
|
BRS1, BRI1 SUPPRESSOR 1, SCPL24, SERINE CARBOXYPEPTIDASE 24 PRECURSOR |
-0.762 |
-0.066 |
| 432 |
255719_at |
AT1G32080
|
AtLrgB, LrgB |
-0.761 |
-0.004 |
| 433 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
-0.761 |
-0.032 |
| 434 |
248246_at |
AT5G53200
|
TRY, TRIPTYCHON |
-0.758 |
0.046 |
| 435 |
263612_at |
AT2G16440
|
MCM4, MINICHROMOSOME MAINTENANCE 4 |
-0.755 |
-0.032 |
| 436 |
255460_at |
AT4G02800
|
unknown |
-0.755 |
0.029 |
| 437 |
267010_at |
AT2G39250
|
SNZ, SCHNARCHZAPFEN |
-0.755 |
-0.102 |
| 438 |
263168_at |
AT1G03020
|
[Thioredoxin superfamily protein] |
-0.753 |
-0.009 |
| 439 |
258025_at |
AT3G19480
|
3-PGDH, PGDH3, phosphoglycerate dehydrogenase 3 |
-0.753 |
-0.071 |
| 440 |
266655_at |
AT2G25880
|
AtAUR2, ataurora2, AUR2, ataurora2 |
-0.753 |
-0.076 |
| 441 |
265722_at |
AT2G40100
|
LHCB4.3, light harvesting complex photosystem II |
-0.752 |
-0.021 |
| 442 |
261907_at |
AT1G65060
|
4CL3, 4-coumarate:CoA ligase 3 |
-0.751 |
-0.074 |
| 443 |
245876_at |
AT1G26230
|
Cpn60beta4, chaperonin-60beta4 |
-0.747 |
-0.039 |
| 444 |
258003_at |
AT3G29030
|
ATEXP5, ARABIDOPSIS THALIANA EXPANSIN 5, ATEXPA5, ARABIDOPSIS THALIANA EXPANSIN A5, ATHEXP ALPHA 1.4, EXP5, EXPANSIN 5, EXPA5, expansin A5 |
-0.747 |
-0.196 |
| 445 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
-0.747 |
-0.149 |
| 446 |
255284_at |
AT4G04610
|
APR, APR1, APS reductase 1, ATAPR1, PRH19, PAPS REDUCTASE HOMOLOG 19 |
-0.745 |
0.012 |
| 447 |
248891_at |
AT5G46280
|
MCM3, MINICHROMOSOME MAINTENANCE 3 |
-0.744 |
-0.028 |
| 448 |
247268_at |
AT5G64080
|
AtXYP1, XYP1, xylogen protein 1 |
-0.742 |
0.001 |
| 449 |
245343_at |
AT4G15830
|
[ARM repeat superfamily protein] |
-0.742 |
0.023 |
| 450 |
256673_at |
AT3G52370
|
FLA15, FASCICLIN-like arabinogalactan protein 15 precursor |
-0.739 |
0.013 |
| 451 |
260146_at |
AT1G52770
|
[Phototropic-responsive NPH3 family protein] |
-0.738 |
0.001 |
| 452 |
248963_at |
AT5G45700
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.735 |
-0.031 |
| 453 |
265305_at |
AT2G20340
|
AAS, aromatic aldehyde synthase, AtAAS |
-0.733 |
-0.069 |
| 454 |
266384_at |
AT2G14660
|
unknown |
-0.733 |
0.081 |
| 455 |
258386_at |
AT3G15520
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.729 |
-0.043 |
| 456 |
251309_at |
AT3G61220
|
SDR1, short-chain dehydrogenase/reductase 1 |
-0.729 |
-0.006 |
| 457 |
250434_at |
AT5G10390
|
H3.1, histone 3.1 |
-0.727 |
0.035 |
| 458 |
266089_at |
AT2G38010
|
[Neutral/alkaline non-lysosomal ceramidase] |
-0.726 |
0.041 |
| 459 |
258480_at |
AT3G02640
|
unknown |
-0.726 |
-0.012 |
| 460 |
245748_at |
AT1G51140
|
AKS1, ABA-responsive kinase substrate 1, FBH3, FLOWERING BHLH 3 |
-0.725 |
-0.178 |
| 461 |
254125_at |
AT4G24670
|
TAR2, tryptophan aminotransferase related 2 |
-0.722 |
0.031 |
| 462 |
255450_at |
AT4G02850
|
[phenazine biosynthesis PhzC/PhzF family protein] |
-0.721 |
-0.017 |
| 463 |
252888_at |
AT4G39210
|
APL3 |
-0.720 |
0.034 |
| 464 |
260388_at |
AT1G74070
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.719 |
-0.076 |
| 465 |
258589_at |
AT3G04290
|
ATLTL1, LTL1, Li-tolerant lipase 1 |
-0.719 |
-0.029 |
| 466 |
259681_at |
AT1G77760
|
GNR1, NIA1, nitrate reductase 1, NR1, NITRATE REDUCTASE 1 |
-0.718 |
0.034 |
| 467 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
-0.717 |
0.116 |
| 468 |
249425_at |
AT5G39790
|
5'-AMP-activated protein kinase-related |
-0.716 |
-0.041 |
| 469 |
255438_at |
AT4G03070
|
AOP, AOP1, AOP1.1 |
-0.716 |
-0.102 |
| 470 |
263034_at |
AT1G24020
|
MLP423, MLP-like protein 423 |
-0.714 |
0.007 |
| 471 |
248921_at |
AT5G45950
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.713 |
-0.084 |
| 472 |
249472_at |
AT5G39210
|
CRR7, CHLORORESPIRATORY REDUCTION 7 |
-0.713 |
-0.031 |
| 473 |
263845_at |
AT2G37040
|
ATPAL1, PAL1, PHE ammonia lyase 1 |
-0.711 |
0.015 |
| 474 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
-0.710 |
-0.009 |
| 475 |
264014_at |
AT2G21210
|
SAUR6, SMALL AUXIN UPREGULATED RNA 6 |
-0.710 |
-0.030 |
| 476 |
245049_at |
ATCG00050
|
RPS16, ribosomal protein S16 |
-0.707 |
0.021 |
| 477 |
246021_at |
AT5G21100
|
[Plant L-ascorbate oxidase] |
-0.707 |
-0.046 |
| 478 |
259432_at |
AT1G01520
|
ASG4, ALTERED SEED GERMINATION 4 |
-0.707 |
0.061 |
| 479 |
247400_at |
AT5G62840
|
[Phosphoglycerate mutase family protein] |
-0.706 |
-0.008 |
| 480 |
255877_at |
AT2G40460
|
[Major facilitator superfamily protein] |
-0.704 |
0.066 |
| 481 |
252853_at |
AT4G39710
|
FKBP16-2, FK506-binding protein 16-2, PnsL4, Photosynthetic NDH subcomplex L 4 |
-0.704 |
-0.016 |
| 482 |
250633_at |
AT5G07460
|
ATMSRA2, ARABIDOPSIS THALIANA METHIONINE SULFOXIDE REDUCTASE 2, PMSR2, peptidemethionine sulfoxide reductase 2 |
-0.701 |
0.003 |
| 483 |
266329_at |
AT2G01590
|
CRR3, CHLORORESPIRATORY REDUCTION 3 |
-0.700 |
-0.002 |
| 484 |
256983_at |
AT3G13470
|
Cpn60beta2, chaperonin-60beta2 |
-0.700 |
0.007 |
| 485 |
251920_at |
AT3G53900
|
PYRR, PYRIMIDINE R, UPP, uracil phosphoribosyltransferase |
-0.699 |
0.012 |
| 486 |
253835_at |
AT4G27820
|
BGLU9, beta glucosidase 9 |
-0.699 |
0.040 |
| 487 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-0.699 |
-0.050 |
| 488 |
257954_at |
AT3G21760
|
HYR1, HYPOSTATIN RESISTANCE 1 |
-0.698 |
-0.052 |
| 489 |
245213_at |
AT1G44575
|
CP22, NPQ4, NONPHOTOCHEMICAL QUENCHING 4, PSBS, PHOTOSYSTEM II SUBUNIT S |
-0.698 |
0.031 |
| 490 |
264262_at |
AT1G09200
|
H3.1, histone 3.1 |
-0.695 |
0.000 |
| 491 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
-0.694 |
0.021 |
| 492 |
246986_at |
AT5G67280
|
RLK, receptor-like kinase |
-0.691 |
-0.094 |
| 493 |
267549_at |
AT2G32640
|
[Lycopene beta/epsilon cyclase protein] |
-0.687 |
-0.052 |
| 494 |
254683_at |
AT4G13800
|
unknown |
-0.684 |
0.039 |
| 495 |
255645_at |
AT4G00880
|
SAUR31, SMALL AUXIN UPREGULATED RNA 31 |
-0.684 |
-0.051 |
| 496 |
266253_at |
AT2G27840
|
HD2D, histone deacetylase 2D, HDA13, HISTONE DEACETYLASE 13, HDT04, HDT4 |
-0.682 |
0.044 |
| 497 |
247716_at |
AT5G59350
|
unknown |
-0.682 |
-0.094 |
| 498 |
252482_at |
AT3G46670
|
UGT76E11, UDP-glucosyl transferase 76E11 |
-0.682 |
-0.078 |
| 499 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
-0.682 |
0.012 |
| 500 |
247415_at |
AT5G63060
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.681 |
-0.026 |
| 501 |
248075_at |
AT5G55740
|
CRR21, chlororespiratory reduction 21 |
-0.681 |
-0.136 |
| 502 |
248330_at |
AT5G52810
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.681 |
-0.039 |
| 503 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
-0.676 |
-0.009 |
| 504 |
258719_at |
AT3G09540
|
[Pectin lyase-like superfamily protein] |
-0.676 |
-0.054 |
| 505 |
249278_at |
AT5G41900
|
[alpha/beta-Hydrolases superfamily protein] |
-0.675 |
-0.005 |
| 506 |
247977_at |
AT5G56850
|
unknown |
-0.675 |
-0.166 |
| 507 |
249410_at |
AT5G40380
|
CRK42, cysteine-rich RLK (RECEPTOR-like protein kinase) 42 |
-0.675 |
-0.120 |
| 508 |
255410_at |
AT4G03100
|
[Rho GTPase activating protein with PAK-box/P21-Rho-binding domain] |
-0.674 |
-0.028 |
| 509 |
253966_at |
AT4G26520
|
AtFBA7, FBA7, fructose-bisphosphate aldolase 7 |
-0.672 |
-0.082 |
| 510 |
253753_at |
AT4G29030
|
[Putative membrane lipoprotein] |
-0.671 |
-0.005 |
| 511 |
245304_at |
AT4G15630
|
[Uncharacterised protein family (UPF0497)] |
-0.670 |
-0.086 |
| 512 |
251509_at |
AT3G59010
|
PME35, pectin methylesterase 35, PME61, pectin methylesterase 61 |
-0.667 |
0.019 |
| 513 |
253790_at |
AT4G28660
|
PSB28, photosystem II reaction center PSB28 protein |
-0.667 |
0.010 |
| 514 |
248139_at |
AT5G54970
|
unknown |
-0.665 |
-0.001 |
| 515 |
257628_at |
AT3G26290
|
CYP71B26, cytochrome P450, family 71, subfamily B, polypeptide 26 |
-0.664 |
-0.031 |
| 516 |
249932_at |
AT5G22390
|
unknown |
-0.663 |
0.021 |
| 517 |
251727_at |
AT3G56290
|
unknown |
-0.662 |
-0.027 |
| 518 |
259878_at |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
-0.662 |
0.006 |
| 519 |
247549_at |
AT5G61420
|
AtMYB28, HAG1, HIGH ALIPHATIC GLUCOSINOLATE 1, MYB28, myb domain protein 28, PMG1, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 1 |
-0.661 |
-0.045 |
| 520 |
253088_at |
AT4G36220
|
CYP84A1, CYTOCHROME P450 84A1, FAH1, ferulic acid 5-hydroxylase 1 |
-0.659 |
-0.039 |
| 521 |
262376_at |
AT1G72970
|
EDA17, embryo sac development arrest 17, HTH, HOTHEAD |
-0.659 |
-0.051 |
| 522 |
253411_at |
AT4G32980
|
ATH1, homeobox gene 1 |
-0.658 |
-0.133 |
| 523 |
267057_at |
AT2G32500
|
[Stress responsive alpha-beta barrel domain protein] |
-0.658 |
-0.111 |
| 524 |
262212_at |
AT1G74890
|
ARR15, response regulator 15 |
-0.657 |
0.038 |
| 525 |
256441_at |
AT3G10940
|
LSF2, LIKE SEX4 2 |
-0.657 |
-0.137 |
| 526 |
251142_at |
AT5G01015
|
unknown |
-0.655 |
0.010 |
| 527 |
258470_at |
AT3G06035
|
[Glycoprotein membrane precursor GPI-anchored] |
-0.654 |
-0.023 |
| 528 |
258895_at |
AT3G05600
|
[alpha/beta-Hydrolases superfamily protein] |
-0.653 |
-0.146 |
| 529 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
-0.650 |
-0.054 |
| 530 |
253598_at |
AT4G30800
|
[Nucleic acid-binding, OB-fold-like protein] |
-0.650 |
-0.021 |
| 531 |
256655_at |
AT3G18890
|
AtTic62, translocon at the inner envelope membrane of chloroplasts 62, Tic62, translocon at the inner envelope membrane of chloroplasts 62 |
-0.649 |
-0.045 |
| 532 |
261570_at |
AT1G01120
|
KCS1, 3-ketoacyl-CoA synthase 1 |
-0.646 |
-0.018 |
| 533 |
266291_at |
AT2G29320
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.645 |
0.002 |
| 534 |
250968_at |
AT5G02890
|
[HXXXD-type acyl-transferase family protein] |
-0.642 |
-0.018 |
| 535 |
247040_at |
AT5G67150
|
[HXXXD-type acyl-transferase family protein] |
-0.642 |
-0.035 |
| 536 |
262526_at |
AT1G17050
|
AtSPS2, SPS2, solanesyl diphosphate synthase 2 |
-0.641 |
-0.034 |
| 537 |
250696_at |
AT5G06790
|
unknown |
-0.641 |
0.032 |
| 538 |
249120_at |
AT5G43750
|
NDH18, NAD(P)H dehydrogenase 18, PnsB5, Photosynthetic NDH subcomplex B 5 |
-0.638 |
0.003 |
| 539 |
247486_at |
AT5G62140
|
unknown |
-0.638 |
-0.111 |
| 540 |
253040_at |
AT4G37800
|
XTH7, xyloglucan endotransglucosylase/hydrolase 7 |
-0.637 |
0.031 |
| 541 |
245165_at |
AT2G33180
|
unknown |
-0.637 |
-0.045 |
| 542 |
247235_at |
AT5G64580
|
EMB3144, EMBRYO DEFECTIVE 3144 |
-0.637 |
-0.024 |
| 543 |
253875_at |
AT4G27520
|
AtENODL2, ENODL2, early nodulin-like protein 2 |
-0.635 |
-0.004 |
| 544 |
253480_at |
AT4G31840
|
AtENODL15, ENODL15, early nodulin-like protein 15 |
-0.632 |
0.009 |
| 545 |
255513_at |
AT4G02060
|
MCM7, PRL, PROLIFERA |
-0.631 |
-0.029 |
| 546 |
245524_at |
AT4G15920
|
AtSWEET17, SWEET17 |
-0.631 |
0.040 |
| 547 |
263452_at |
AT2G22190
|
TPPE, trehalose-6-phosphate phosphatase E |
-0.631 |
-0.065 |
| 548 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
-0.630 |
0.034 |
| 549 |
252358_at |
AT3G48425
|
[DNAse I-like superfamily protein] |
-0.630 |
0.024 |
| 550 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
-0.628 |
-0.094 |
| 551 |
252148_at |
AT3G51280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.628 |
-0.046 |
| 552 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
-0.626 |
0.024 |
| 553 |
250502_at |
AT5G09590
|
HSC70-5, HEAT SHOCK COGNATE, MTHSC70-2, mitochondrial HSO70 2 |
-0.626 |
-0.000 |
| 554 |
266413_at |
AT2G38740
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.625 |
0.013 |
| 555 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
-0.625 |
-0.103 |
| 556 |
261660_at |
AT1G18370
|
ATNACK1, ARABIDOPSIS NPK1-ACTIVATING KINESIN 1, HIK, HINKEL, NACK1, NPK1-ACTIVATING KINESIN 1 |
-0.624 |
0.004 |
| 557 |
265991_at |
AT2G24120
|
PDE319, PIGMENT DEFECTIVE 319, SCA3, SCABRA 3 |
-0.624 |
-0.013 |
| 558 |
263474_at |
AT2G31725
|
unknown |
-0.623 |
0.037 |
| 559 |
256585_at |
AT3G28760
|
unknown |
-0.621 |
-0.022 |
| 560 |
252366_at |
AT3G48420
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.619 |
-0.023 |
| 561 |
251730_at |
AT3G56330
|
[N2,N2-dimethylguanosine tRNA methyltransferase] |
-0.619 |
-0.002 |
| 562 |
261420_at |
AT1G07720
|
KCS3, 3-ketoacyl-CoA synthase 3 |
-0.618 |
-0.109 |
| 563 |
252478_at |
AT3G46540
|
[ENTH/VHS family protein] |
-0.617 |
-0.057 |
| 564 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
-0.617 |
-0.084 |
| 565 |
263483_at |
AT2G04030
|
AtHsp90.5, HEAT SHOCK PROTEIN 90.5, AtHsp90C, CR88, EMB1956, EMBRYO DEFECTIVE 1956, Hsp88.1, HEAT SHOCK PROTEIN 88.1, HSP90.5, HEAT SHOCK PROTEIN 90.5 |
-0.616 |
-0.002 |
| 566 |
258409_at |
AT3G17640
|
[Leucine-rich repeat (LRR) family protein] |
-0.616 |
-0.047 |
| 567 |
257801_at |
AT3G18750
|
ATWNK6, ARABIDOPSIS THALIANA WITH NO K 6, WNK6, with no lysine (K) kinase 6, ZIK5 |
-0.616 |
-0.081 |
| 568 |
263882_at |
AT2G21790
|
ATRNR1, RIBONUCLEOTIDE REDUCTASE LARGE SUBUNIT 1, CLS8, CRINKLY LEAVES 8, DPD2, defective in pollen organelle DNA degradation 2, R1, RIBONUCLEOTIDE REDUCTASE 1, RNR1, ribonucleotide reductase 1 |
-0.615 |
-0.039 |
| 569 |
251119_at |
AT3G63510
|
[FMN-linked oxidoreductases superfamily protein] |
-0.615 |
-0.044 |
| 570 |
250529_at |
AT5G08610
|
PDE340, PIGMENT DEFECTIVE 340 |
-0.613 |
0.014 |
| 571 |
257891_at |
AT3G17170
|
RFC3, REGULATOR OF FATTY-ACID COMPOSITION 3 |
-0.613 |
-0.000 |
| 572 |
264319_at |
AT1G04110
|
AtSDD1, SDD1, STOMATAL DENSITY AND DISTRIBUTION 1 |
-0.610 |
0.046 |
| 573 |
255676_at |
AT4G00490
|
BAM2, beta-amylase 2, BMY9, BETA-AMYLASE 9 |
-0.610 |
0.016 |
| 574 |
253566_at |
AT4G31210
|
[DNA topoisomerase, type IA, core] |
-0.610 |
-0.027 |
| 575 |
265097_at |
AT1G04020
|
ATBARD1, BARD1, breast cancer associated RING 1, ROW1 |
-0.607 |
0.069 |
| 576 |
247651_at |
AT5G59870
|
HTA6, histone H2A 6 |
-0.607 |
0.003 |
| 577 |
247417_at |
AT5G63040
|
unknown |
-0.606 |
0.034 |
| 578 |
252442_at |
AT3G46940
|
DUT1, DUTP-PYROPHOSPHATASE-LIKE 1 |
-0.606 |
0.009 |
| 579 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
-0.606 |
0.014 |
| 580 |
249813_at |
AT5G23940
|
DCR, DEFECTIVE IN CUTICULAR RIDGES, EMB3009, EMBRYO DEFECTIVE 3009, PEL3, PERMEABLE LEAVES3 |
-0.605 |
-0.129 |
| 581 |
264783_at |
AT1G08650
|
ATPPCK1, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 1, PPCK1, phosphoenolpyruvate carboxylase kinase 1 |
-0.605 |
-0.164 |
| 582 |
265939_at |
AT2G19650
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.605 |
-0.055 |
| 583 |
247109_at |
AT5G65870
|
ATPSK5, phytosulfokine 5 precursor, PSK5, PSK5, phytosulfokine 5 precursor |
-0.605 |
-0.128 |
| 584 |
256302_at |
AT1G69526
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.604 |
-0.071 |
| 585 |
263031_at |
AT1G24070
|
ATCSLA10, cellulose synthase-like A10, CSLA10, CELLULOSE SYNTHASE LIKE A10, CSLA10, cellulose synthase-like A10 |
-0.604 |
0.075 |
| 586 |
260466_at |
AT1G10900
|
[Phosphatidylinositol-4-phosphate 5-kinase family protein] |
-0.604 |
-0.072 |
| 587 |
264511_at |
AT1G09350
|
AtGolS3, galactinol synthase 3, GolS3, galactinol synthase 3 |
-0.603 |
-0.027 |
| 588 |
260077_at |
AT1G73620
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.602 |
-0.018 |
| 589 |
248094_at |
AT5G55220
|
[trigger factor type chaperone family protein] |
-0.601 |
-0.013 |
| 590 |
266418_at |
AT2G38750
|
ANNAT4, annexin 4, AtANN4 |
-0.598 |
-0.006 |
| 591 |
255549_at |
AT4G01950
|
ATGPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3, GPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3 |
-0.598 |
-0.015 |
| 592 |
245676_at |
AT1G56670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.598 |
-0.182 |
| 593 |
255622_at |
AT4G01070
|
GT72B1, UGT72B1, UDP-GLUCOSE-DEPENDENT GLUCOSYLTRANSFERASE 72 B1 |
-0.597 |
-0.153 |
| 594 |
252619_at |
AT3G45210
|
unknown |
-0.597 |
-0.009 |
| 595 |
266447_at |
AT2G43290
|
MSS3, multicopy suppressors of snf4 deficiency in yeast 3 |
-0.596 |
-0.089 |
| 596 |
249138_at |
AT5G43070
|
WPP1, WPP domain protein 1 |
-0.595 |
-0.198 |
| 597 |
261804_at |
AT1G30530
|
UGT78D1, UDP-glucosyl transferase 78D1 |
-0.595 |
-0.055 |
| 598 |
257474_at |
AT1G80850
|
[DNA glycosylase superfamily protein] |
-0.595 |
-0.078 |
| 599 |
257173_at |
AT3G23810
|
ATSAHH2, S-ADENOSYL-L-HOMOCYSTEINE (SAH) HYDROLASE 2, SAHH2, S-adenosyl-l-homocysteine (SAH) hydrolase 2 |
-0.595 |
0.025 |
| 600 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
-0.592 |
0.022 |