|
probeID |
AGICode |
Annotation |
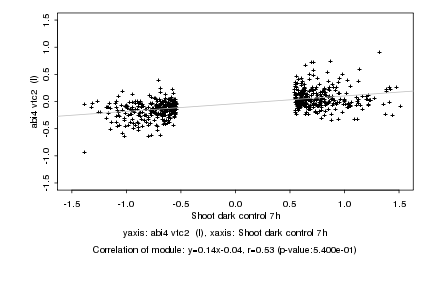
Log2 signal ratio
Shoot dark control 7h |
Log2 signal ratio
abi4 vtc2 (I) |
| 1 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
1.510 |
-0.091 |
| 2 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
1.474 |
0.273 |
| 3 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
1.434 |
-0.255 |
| 4 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
1.417 |
0.226 |
| 5 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
1.404 |
0.273 |
| 6 |
260668_at |
AT1G19530
|
unknown |
1.404 |
-0.018 |
| 7 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
1.383 |
0.198 |
| 8 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
1.381 |
0.238 |
| 9 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
1.370 |
-0.232 |
| 10 |
258383_at |
AT3G15440
|
unknown |
1.351 |
-0.046 |
| 11 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
1.321 |
0.909 |
| 12 |
260527_at |
AT2G47270
|
UPB1, UPBEAT1 |
1.267 |
0.056 |
| 13 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
1.238 |
0.025 |
| 14 |
254001_at |
AT4G26260
|
MIOX4, myo-inositol oxygenase 4 |
1.235 |
0.026 |
| 15 |
258327_at |
AT3G22640
|
PAP85 |
1.226 |
0.021 |
| 16 |
250666_at |
AT5G07100
|
WRKY26, WRKY DNA-binding protein 26 |
1.217 |
0.116 |
| 17 |
256743_at |
AT3G29370
|
P1R3, P1R3 |
1.215 |
-0.061 |
| 18 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
1.214 |
-0.049 |
| 19 |
251012_at |
AT5G02580
|
unknown |
1.210 |
0.051 |
| 20 |
253322_at |
AT4G33980
|
unknown |
1.196 |
0.039 |
| 21 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
1.193 |
0.097 |
| 22 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
1.163 |
0.103 |
| 23 |
249454_at |
AT5G39520
|
unknown |
1.159 |
-0.143 |
| 24 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
1.143 |
-0.061 |
| 25 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
1.134 |
0.598 |
| 26 |
247919_at |
AT5G57650
|
[eukaryotic translation initiation factor-related] |
1.123 |
0.041 |
| 27 |
247954_at |
AT5G56870
|
BGAL4, beta-galactosidase 4 |
1.122 |
0.381 |
| 28 |
250182_at |
AT5G14470
|
AtGALK2, GALK2, galactokinase 2 |
1.121 |
-0.021 |
| 29 |
258005_at |
AT3G19390
|
[Granulin repeat cysteine protease family protein] |
1.118 |
0.003 |
| 30 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
1.118 |
-0.331 |
| 31 |
267036_at |
AT2G38465
|
unknown |
1.105 |
0.081 |
| 32 |
260662_at |
AT1G19540
|
[NmrA-like negative transcriptional regulator family protein] |
1.101 |
-0.060 |
| 33 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
1.085 |
-0.324 |
| 34 |
253373_at |
AT4G33150
|
LKR, LKR/SDH, LYSINE-KETOGLUTARATE REDUCTASE/SACCHAROPINE DEHYDROGENASE, SDH, SACCHAROPINE DEHYDROGENASE |
1.064 |
-0.003 |
| 35 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
1.055 |
-0.062 |
| 36 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
1.054 |
-0.075 |
| 37 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
1.047 |
0.278 |
| 38 |
245233_at |
AT4G25580
|
[CAP160 protein] |
1.033 |
-0.030 |
| 39 |
262580_at |
AT1G15330
|
[Cystathionine beta-synthase (CBS) protein] |
1.031 |
-0.054 |
| 40 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
1.030 |
-0.101 |
| 41 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
1.023 |
0.388 |
| 42 |
252539_at |
AT3G45730
|
unknown |
1.020 |
-0.104 |
| 43 |
258225_at |
AT3G15630
|
unknown |
1.011 |
0.159 |
| 44 |
246251_at |
AT4G37220
|
[Cold acclimation protein WCOR413 family] |
1.001 |
-0.045 |
| 45 |
257670_at |
AT3G20340
|
unknown |
0.999 |
0.029 |
| 46 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
0.997 |
0.052 |
| 47 |
253829_at |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
0.991 |
0.008 |
| 48 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
0.987 |
-0.042 |
| 49 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
0.981 |
-0.196 |
| 50 |
246929_at |
AT5G25210
|
unknown |
0.977 |
0.503 |
| 51 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
0.957 |
0.041 |
| 52 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
0.951 |
0.163 |
| 53 |
251039_at |
AT5G02020
|
SIS, Salt Induced Serine rich |
0.951 |
-0.018 |
| 54 |
264580_at |
AT1G05340
|
unknown |
0.947 |
0.428 |
| 55 |
264467_at |
AT1G10140
|
[Uncharacterised conserved protein UCP031279] |
0.944 |
0.151 |
| 56 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
0.940 |
0.367 |
| 57 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
0.937 |
-0.318 |
| 58 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
0.932 |
-0.010 |
| 59 |
259244_at |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
0.925 |
-0.043 |
| 60 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
0.923 |
0.260 |
| 61 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
0.917 |
-0.141 |
| 62 |
265539_at |
AT2G15830
|
unknown |
0.915 |
0.203 |
| 63 |
260741_at |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
0.914 |
-0.027 |
| 64 |
251026_at |
AT5G02200
|
FHL, far-red-elongated hypocotyl1-like |
0.908 |
0.031 |
| 65 |
265536_at |
AT2G15880
|
[Leucine-rich repeat (LRR) family protein] |
0.895 |
-0.023 |
| 66 |
246238_at |
AT4G36670
|
AtPLT6, AtPMT6, PLT6, polyol transporter 6, PMT6, polyol/monosaccharide transporter 6 |
0.891 |
0.261 |
| 67 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
0.888 |
-0.068 |
| 68 |
252570_at |
AT3G45300
|
ATIVD, IVD, isovaleryl-CoA-dehydrogenase, IVDH, ISOVALERYL-COA-DEHYDROGENASE |
0.887 |
0.038 |
| 69 |
259982_at |
AT1G76410
|
ATL8 |
0.886 |
0.068 |
| 70 |
254197_at |
AT4G24040
|
ATTRE1, TRE1, trehalase 1 |
0.883 |
0.097 |
| 71 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
0.881 |
0.326 |
| 72 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
0.879 |
-0.231 |
| 73 |
264238_at |
AT1G54740
|
unknown |
0.877 |
-0.345 |
| 74 |
252924_at |
AT4G39070
|
BBX20, B-box domain protein 20, BZS1, BZS1 |
0.875 |
0.054 |
| 75 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
0.871 |
0.747 |
| 76 |
260748_at |
AT1G49210
|
[RING/U-box superfamily protein] |
0.859 |
0.018 |
| 77 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
0.859 |
0.213 |
| 78 |
255381_at |
AT4G03510
|
ATRMA1, RMA1, RING membrane-anchor 1 |
0.858 |
0.264 |
| 79 |
256061_at |
AT1G07040
|
unknown |
0.857 |
0.109 |
| 80 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.853 |
-0.129 |
| 81 |
263118_at |
AT1G03090
|
MCCA |
0.853 |
-0.024 |
| 82 |
250649_at |
AT5G06690
|
WCRKC1, WCRKC thioredoxin 1 |
0.852 |
0.386 |
| 83 |
264246_at |
AT1G60140
|
ATTPS10, trehalose phosphate synthase, TPS10, trehalose phosphate synthase, TPS10, TREHALOSE PHOSPHATE SYNTHASE 10 |
0.852 |
0.239 |
| 84 |
248279_at |
AT5G52910
|
ATIM, TIMELESS |
0.851 |
-0.000 |
| 85 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
0.847 |
-0.016 |
| 86 |
265266_at |
AT2G42890
|
AML2, MEI2-like 2, ML2, MEI2-like 2 |
0.845 |
0.078 |
| 87 |
246831_at |
AT5G26340
|
ATSTP13, SUGAR TRANSPORT PROTEIN 13, MSS1, STP13, SUGAR TRANSPORT PROTEIN 13 |
0.843 |
0.540 |
| 88 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
0.833 |
0.128 |
| 89 |
266477_at |
AT2G31085
|
AtCLE6, CLE6, CLAVATA3/ESR-RELATED 6 |
0.827 |
-0.041 |
| 90 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
0.824 |
-0.055 |
| 91 |
256965_at |
AT3G13450
|
DIN4, DARK INDUCIBLE 4 |
0.823 |
0.117 |
| 92 |
249869_at |
AT5G23050
|
AAE17, acyl-activating enzyme 17 |
0.822 |
0.169 |
| 93 |
266259_at |
AT2G27830
|
unknown |
0.821 |
-0.003 |
| 94 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
0.818 |
-0.171 |
| 95 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
0.815 |
0.051 |
| 96 |
265481_at |
AT2G15960
|
unknown |
0.815 |
0.141 |
| 97 |
247474_at |
AT5G62280
|
unknown |
0.814 |
-0.007 |
| 98 |
260688_at |
AT1G17665
|
unknown |
0.809 |
0.011 |
| 99 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
0.809 |
0.238 |
| 100 |
263881_at |
AT2G21820
|
unknown |
0.809 |
-0.243 |
| 101 |
247013_at |
AT5G67480
|
ATBT4, BT4, BTB and TAZ domain protein 4 |
0.808 |
0.246 |
| 102 |
258939_at |
AT3G10020
|
unknown |
0.800 |
0.193 |
| 103 |
246114_at |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
0.796 |
0.061 |
| 104 |
252993_at |
AT4G38540
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
0.796 |
0.167 |
| 105 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
0.790 |
0.191 |
| 106 |
264482_at |
AT1G77210
|
AtSTP14, sugar transport protein 14, STP14, sugar transport protein 14 |
0.787 |
0.096 |
| 107 |
263653_at |
AT1G04310
|
ERS2, ethylene response sensor 2 |
0.786 |
-0.067 |
| 108 |
254193_at |
AT4G23870
|
unknown |
0.786 |
0.016 |
| 109 |
258395_at |
AT3G15500
|
ANAC055, NAC domain containing protein 55, ATNAC3, NAC domain containing protein 3, NAC055, NAC domain containing protein 55, NAC3, NAC domain containing protein 3 |
0.784 |
0.050 |
| 110 |
259058_at |
AT3G03470
|
CYP89A9, cytochrome P450, family 87, subfamily A, polypeptide 9 |
0.783 |
0.071 |
| 111 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
0.781 |
-0.101 |
| 112 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
0.781 |
-0.296 |
| 113 |
251356_at |
AT3G61060
|
AtPP2-A13, phloem protein 2-A13, PP2-A13, phloem protein 2-A13 |
0.780 |
-0.153 |
| 114 |
261211_at |
AT1G12780
|
ATUGE1, A. THALIANA UDP-GLC 4-EPIMERASE 1, UGE1, UDP-D-glucose/UDP-D-galactose 4-epimerase 1 |
0.779 |
0.135 |
| 115 |
245440_at |
AT4G16680
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.776 |
-0.032 |
| 116 |
263475_at |
AT2G31945
|
unknown |
0.776 |
0.181 |
| 117 |
248193_at |
AT5G54080
|
AtHGO, HGO, homogentisate 1,2-dioxygenase |
0.772 |
0.029 |
| 118 |
248969_at |
AT5G45310
|
unknown |
0.769 |
-0.173 |
| 119 |
264551_at |
AT1G09460
|
[Carbohydrate-binding X8 domain superfamily protein] |
0.768 |
-0.054 |
| 120 |
264045_at |
AT2G22450
|
AtRIBA2, RIBA2, homolog of ribA 2 |
0.764 |
0.326 |
| 121 |
263281_at |
AT2G14160
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.763 |
-0.011 |
| 122 |
258227_at |
AT3G15620
|
UVR3, UV REPAIR DEFECTIVE 3 |
0.757 |
-0.082 |
| 123 |
264124_at |
AT1G79360
|
ATOCT2, organic cation/carnitine transporter 2, OCT2, ORGANIC CATION TRANSPORTER 2, OCT2, organic cation/carnitine transporter 2 |
0.751 |
-0.074 |
| 124 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
0.751 |
0.432 |
| 125 |
248029_at |
AT5G55700
|
BAM4, beta-amylase 4, BMY6, BETA-AMYLASE 6 |
0.747 |
0.082 |
| 126 |
256697_at |
AT3G20660
|
AtOCT4, organic cation/carnitine transporter4, OCT4, organic cation/carnitine transporter4 |
0.746 |
0.122 |
| 127 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.745 |
-0.211 |
| 128 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
0.744 |
0.043 |
| 129 |
255742_at |
AT1G25560
|
AtTEM1, EDF1, ETHYLENE RESPONSE DNA BINDING FACTOR 1, TEM1, TEMPRANILLO 1 |
0.743 |
0.156 |
| 130 |
251647_at |
AT3G57770
|
[Protein kinase superfamily protein] |
0.740 |
-0.053 |
| 131 |
261272_at |
AT1G26665
|
[Mediator complex, subunit Med10] |
0.740 |
0.073 |
| 132 |
250940_at |
AT5G03310
|
SAUR44, SMALL AUXIN UPREGULATED RNA 44 |
0.740 |
0.024 |
| 133 |
255543_at |
AT4G01870
|
[tolB protein-related] |
0.735 |
0.268 |
| 134 |
247835_at |
AT5G57910
|
unknown |
0.735 |
0.059 |
| 135 |
261205_at |
AT1G12790
|
unknown |
0.734 |
0.044 |
| 136 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.733 |
-0.169 |
| 137 |
260011_at |
AT1G68110
|
[ENTH/ANTH/VHS superfamily protein] |
0.729 |
-0.202 |
| 138 |
248895_at |
AT5G46330
|
FLS2, FLAGELLIN-SENSITIVE 2 |
0.728 |
0.228 |
| 139 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.726 |
0.223 |
| 140 |
247128_at |
AT5G66110
|
HIPP27, heavy metal associated isoprenylated plant protein 27 |
0.726 |
-0.053 |
| 141 |
252109_at |
AT3G51540
|
unknown |
0.721 |
-0.054 |
| 142 |
253737_at |
AT4G28703
|
[RmlC-like cupins superfamily protein] |
0.716 |
0.023 |
| 143 |
245794_at |
AT1G32170
|
XTH30, xyloglucan endotransglucosylase/hydrolase 30, XTR4, xyloglucan endotransglycosylase 4 |
0.713 |
-0.033 |
| 144 |
246028_at |
AT5G21170
|
5'-AMP-activated protein kinase beta-2 subunit protein |
0.711 |
-0.058 |
| 145 |
254954_at |
AT4G10910
|
unknown |
0.711 |
-0.039 |
| 146 |
245317_at |
AT4G15610
|
[Uncharacterised protein family (UPF0497)] |
0.710 |
0.214 |
| 147 |
252068_at |
AT3G51440
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.710 |
0.577 |
| 148 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
0.709 |
0.735 |
| 149 |
251116_at |
AT3G63470
|
scpl40, serine carboxypeptidase-like 40 |
0.708 |
-0.110 |
| 150 |
258833_at |
AT3G07274
|
|
0.707 |
0.161 |
| 151 |
266839_at |
AT2G25930
|
ELF3, EARLY FLOWERING 3, PYK20 |
0.707 |
0.493 |
| 152 |
247525_at |
AT5G61380
|
APRR1, AtTOC1, TIMING OF CAB EXPRESSION 1, PRR1, PSEUDO-RESPONSE REGULATOR 1, TOC1, TIMING OF CAB EXPRESSION 1 |
0.706 |
0.208 |
| 153 |
262607_at |
AT1G13990
|
unknown |
0.702 |
0.266 |
| 154 |
247924_at |
AT5G57655
|
[xylose isomerase family protein] |
0.694 |
0.005 |
| 155 |
251221_at |
AT3G62550
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.694 |
0.233 |
| 156 |
262981_at |
AT1G75590
|
SAUR52, SMALL AUXIN UPREGULATED RNA 52 |
0.694 |
0.120 |
| 157 |
265070_at |
AT1G55510
|
BCDH BETA1, branched-chain alpha-keto acid decarboxylase E1 beta subunit |
0.693 |
0.048 |
| 158 |
259595_at |
AT1G28050
|
BBX13, B-box domain protein 13 |
0.690 |
0.114 |
| 159 |
252464_at |
AT3G47160
|
[RING/U-box superfamily protein] |
0.689 |
0.724 |
| 160 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.687 |
0.210 |
| 161 |
248371_at |
AT5G51810
|
AT2353, ATGA20OX2, GA20OX2, gibberellin 20 oxidase 2 |
0.684 |
-0.056 |
| 162 |
246935_at |
AT5G25350
|
EBF2, EIN3-binding F box protein 2 |
0.682 |
0.016 |
| 163 |
266711_at |
AT2G46740
|
AtGulLO5, GulLO5, L -gulono-1,4-lactone ( L -GulL) oxidase 5 |
0.680 |
-0.133 |
| 164 |
248744_at |
AT5G48250
|
BBX8, B-box domain protein 8 |
0.679 |
0.193 |
| 165 |
247878_at |
AT5G57760
|
unknown |
0.679 |
-0.100 |
| 166 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
0.678 |
-0.130 |
| 167 |
247394_at |
AT5G62865
|
unknown |
0.677 |
0.036 |
| 168 |
267178_at |
AT2G37750
|
unknown |
0.677 |
0.188 |
| 169 |
246396_at |
AT1G58180
|
ATBCA6, A. THALIANA BETA CARBONIC ANHYDRASE 6, BCA6, beta carbonic anhydrase 6 |
0.675 |
0.026 |
| 170 |
267075_at |
AT2G41070
|
ATBZIP12, DPBF4, EEL, ENHANCED EM LEVEL |
0.675 |
-0.078 |
| 171 |
251789_at |
AT3G55450
|
PBL1, PBS1-like 1 |
0.673 |
0.417 |
| 172 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
0.670 |
0.507 |
| 173 |
250680_at |
AT5G06570
|
[alpha/beta-Hydrolases superfamily protein] |
0.670 |
0.062 |
| 174 |
266392_at |
AT2G41280
|
ATM10, M10 |
0.669 |
0.035 |
| 175 |
265672_at |
AT2G31980
|
AtCYS2, PHYTOCYSTATIN 2, CYS2, PHYTOCYSTATIN 2 |
0.667 |
-0.076 |
| 176 |
253281_at |
AT4G34138
|
UGT73B1, UDP-glucosyl transferase 73B1 |
0.666 |
0.042 |
| 177 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
0.665 |
-0.235 |
| 178 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
0.664 |
-0.129 |
| 179 |
249087_at |
AT5G44210
|
ATERF-9, ERF DOMAIN PROTEIN- 9, ATERF9, ERF DOMAIN PROTEIN 9, ERF9, erf domain protein 9 |
0.655 |
0.107 |
| 180 |
247348_at |
AT5G63810
|
AtBGAL10, Arabidopsis thaliana beta-galactosidase 10, BGAL10, beta-galactosidase 10 |
0.654 |
0.131 |
| 181 |
258339_at |
AT3G16120
|
[Dynein light chain type 1 family protein] |
0.651 |
-0.029 |
| 182 |
250580_at |
AT5G07440
|
GDH2, glutamate dehydrogenase 2 |
0.651 |
0.139 |
| 183 |
261658_at |
AT1G50040
|
unknown |
0.649 |
-0.043 |
| 184 |
259694_at |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
0.645 |
-0.049 |
| 185 |
252323_at |
AT3G48530
|
KING1, SNF1-related protein kinase regulatory subunit gamma 1 |
0.645 |
0.023 |
| 186 |
254665_at |
AT4G18340
|
[Glycosyl hydrolase superfamily protein] |
0.644 |
-0.104 |
| 187 |
264209_at |
AT1G22740
|
ATRABG3B, RAB7, RAB75, RABG3B, RAB GTPase homolog G3B |
0.643 |
-0.192 |
| 188 |
264513_at |
AT1G09420
|
G6PD4, glucose-6-phosphate dehydrogenase 4 |
0.642 |
0.664 |
| 189 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
0.639 |
0.090 |
| 190 |
261004_at |
AT1G26450
|
[Carbohydrate-binding X8 domain superfamily protein] |
0.638 |
-0.000 |
| 191 |
258209_at |
AT3G14060
|
unknown |
0.638 |
0.136 |
| 192 |
266140_at |
AT2G28120
|
[Major facilitator superfamily protein] |
0.638 |
-0.038 |
| 193 |
265511_at |
AT2G05540
|
[Glycine-rich protein family] |
0.638 |
-0.235 |
| 194 |
260536_at |
AT2G43400
|
ETFQO, electron-transfer flavoprotein:ubiquinone oxidoreductase |
0.638 |
-0.033 |
| 195 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
0.637 |
-0.033 |
| 196 |
255283_at |
AT4G04620
|
ATG8B, autophagy 8b |
0.636 |
0.068 |
| 197 |
247674_at |
AT5G59930
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.636 |
-0.037 |
| 198 |
252183_at |
AT3G50740
|
UGT72E1, UDP-glucosyl transferase 72E1 |
0.634 |
0.208 |
| 199 |
248509_at |
AT5G50335
|
unknown |
0.634 |
0.102 |
| 200 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
0.630 |
-0.058 |
| 201 |
251245_at |
AT3G62090
|
PIF6, PHYTOCHROME-INTERACTING FACTOR 6, PIL2, phytochrome interacting factor 3-like 2 |
0.630 |
0.053 |
| 202 |
256114_at |
AT1G16850
|
unknown |
0.629 |
-0.036 |
| 203 |
248502_at |
AT5G50450
|
[HCP-like superfamily protein with MYND-type zinc finger] |
0.629 |
0.065 |
| 204 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
0.628 |
-0.089 |
| 205 |
252751_at |
AT3G43430
|
[RING/U-box superfamily protein] |
0.628 |
0.240 |
| 206 |
251621_at |
AT3G57700
|
[Protein kinase superfamily protein] |
0.628 |
0.267 |
| 207 |
252117_at |
AT3G51430
|
SSL5, STRICTOSIDINE SYNTHASE-LIKE 5, YLS2, YELLOW-LEAF-SPECIFIC GENE 2 |
0.627 |
0.252 |
| 208 |
257830_at |
AT3G26690
|
ATNUDT13, ARABIDOPSIS THALIANA NUDIX HYDROLASE HOMOLOG 13, ATNUDX13, nudix hydrolase homolog 13, NUDX13, nudix hydrolase homolog 13 |
0.626 |
0.317 |
| 209 |
249783_at |
AT5G24270
|
ATSOS3, SALT OVERLY SENSITIVE 3, CBL4, CALCINEURIN B-LIKE PROTEIN 4, SOS3, SALT OVERLY SENSITIVE 3 |
0.626 |
0.148 |
| 210 |
248432_at |
AT5G51390
|
unknown |
0.623 |
-0.067 |
| 211 |
262456_at |
AT1G11260
|
ATSTP1, SUGAR TRANSPORTER 1, STP1, sugar transporter 1 |
0.621 |
0.027 |
| 212 |
256930_at |
AT3G22460
|
OASA2, O-acetylserine (thiol) lyase (OAS-TL) isoform A2 |
0.619 |
0.230 |
| 213 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.619 |
-0.028 |
| 214 |
261586_at |
AT1G01640
|
[BTB/POZ domain-containing protein] |
0.618 |
0.191 |
| 215 |
267628_at |
AT2G42280
|
AKS3, ABA-responsive kinase substrate 3, FBH4, FLOWERING BHLH 4 |
0.617 |
0.146 |
| 216 |
251642_at |
AT3G57520
|
AtSIP2, seed imbibition 2, RS2, raffinose synthase 2, SIP2, seed imbibition 2 |
0.614 |
0.017 |
| 217 |
247374_at |
AT5G63190
|
[MA3 domain-containing protein] |
0.612 |
0.027 |
| 218 |
261615_at |
AT1G33050
|
unknown |
0.612 |
-0.042 |
| 219 |
261901_at |
AT1G80920
|
AtJ8, AtToc12, translocon at the outer envelope membrane of chloroplasts 12, DJC22, DNA J protein C22, J8, Toc12, translocon at the outer envelope membrane of chloroplasts 12 |
0.610 |
-0.047 |
| 220 |
263196_at |
AT1G36070
|
[Transducin/WD40 repeat-like superfamily protein] |
0.609 |
-0.036 |
| 221 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.606 |
0.385 |
| 222 |
245076_at |
AT2G23170
|
GH3.3 |
0.606 |
0.209 |
| 223 |
249990_at |
AT5G18540
|
unknown |
0.606 |
0.140 |
| 224 |
264774_at |
AT1G22890
|
unknown |
0.605 |
-0.063 |
| 225 |
250194_at |
AT5G14550
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
0.604 |
0.349 |
| 226 |
260279_at |
AT1G80420
|
ATXRCC1, XRCC1, homolog of X-ray repair cross complementing 1 |
0.604 |
0.158 |
| 227 |
253827_at |
AT4G28085
|
unknown |
0.602 |
-0.028 |
| 228 |
263914_at |
AT2G36400
|
AtGRF3, growth-regulating factor 3, GRF3, growth-regulating factor 3 |
0.601 |
0.155 |
| 229 |
250900_at |
AT5G03470
|
Protein phosphatase 2A regulatory B subunit family protein |
0.601 |
0.154 |
| 230 |
251022_at |
AT5G02150
|
Fes1C, Fes1C |
0.601 |
0.097 |
| 231 |
250032_at |
AT5G18170
|
GDH1, glutamate dehydrogenase 1 |
0.600 |
0.114 |
| 232 |
261613_at |
AT1G49720
|
ABF1, abscisic acid responsive element-binding factor 1, AtABF1 |
0.599 |
0.434 |
| 233 |
266983_at |
AT2G39400
|
[alpha/beta-Hydrolases superfamily protein] |
0.599 |
0.079 |
| 234 |
263963_at |
AT2G36080
|
ABS2, ABNORMAL SHOOT 2, NGAL1, NGATHA-Like 1 |
0.598 |
0.299 |
| 235 |
253722_at |
AT4G29190
|
AtC3H49, AtOZF2, Arabidopsis thaliana oxidation-related zinc finger 2, AtTZF3, OZF2, oxidation-related zinc finger 2, TZF3, tandem zinc finger 3 |
0.598 |
0.136 |
| 236 |
261674_at |
AT1G18270
|
[ketose-bisphosphate aldolase class-II family protein] |
0.597 |
-0.022 |
| 237 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
0.597 |
0.182 |
| 238 |
262322_at |
AT1G27590
|
unknown |
0.597 |
0.056 |
| 239 |
247951_at |
AT5G57240
|
ORP4C, OSBP(oxysterol binding protein)-related protein 4C |
0.594 |
-0.064 |
| 240 |
264338_at |
AT1G70300
|
KUP6, K+ uptake permease 6 |
0.593 |
0.180 |
| 241 |
253617_at |
AT4G30410
|
[sequence-specific DNA binding transcription factors] |
0.592 |
0.119 |
| 242 |
258880_at |
AT3G06420
|
ATG8H, autophagy 8h |
0.592 |
0.019 |
| 243 |
267527_at |
AT2G45610
|
[alpha/beta-Hydrolases superfamily protein] |
0.592 |
-0.103 |
| 244 |
245543_at |
AT4G15260
|
[UDP-Glycosyltransferase superfamily protein] |
0.591 |
0.033 |
| 245 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
0.590 |
0.060 |
| 246 |
262599_at |
AT1G15350
|
unknown |
0.586 |
-0.055 |
| 247 |
267230_at |
AT2G44080
|
ARL, ARGOS-like |
0.585 |
-0.006 |
| 248 |
266054_at |
AT2G40640
|
[RING/U-box superfamily protein] |
0.585 |
0.030 |
| 249 |
246151_at |
AT5G19950
|
unknown |
0.584 |
0.042 |
| 250 |
259015_at |
AT3G07350
|
unknown |
0.583 |
-0.113 |
| 251 |
266156_at |
AT2G28110
|
FRA8, FRAGILE FIBER 8, IRX7, IRREGULAR XYLEM 7 |
0.582 |
0.139 |
| 252 |
253794_at |
AT4G28720
|
YUC8, YUCCA 8 |
0.581 |
-0.128 |
| 253 |
265902_at |
AT2G25590
|
[Plant Tudor-like protein] |
0.580 |
0.138 |
| 254 |
261567_at |
AT1G33055
|
unknown |
0.579 |
-0.047 |
| 255 |
253880_at |
AT4G27590
|
[Heavy metal transport/detoxification superfamily protein ] |
0.578 |
-0.095 |
| 256 |
255625_at |
AT4G01120
|
ATBZIP54, BASIC REGION/LEUCINE ZIPPER MOTIF 5, GBF2, G-box binding factor 2 |
0.577 |
-0.158 |
| 257 |
257832_at |
AT3G26740
|
CCL, CCR-like |
0.575 |
0.150 |
| 258 |
264508_at |
AT1G09570
|
FHY2, FAR RED ELONGATED HYPOCOTYL 2, FRE1, FAR RED ELONGATED 1, HY8, ELONGATED HYPOCOTYL 8, PHYA, phytochrome A |
0.575 |
-0.013 |
| 259 |
263157_at |
AT1G54100
|
ALDH7B4, aldehyde dehydrogenase 7B4 |
0.574 |
-0.059 |
| 260 |
262977_at |
AT1G75490
|
[Integrase-type DNA-binding superfamily protein] |
0.574 |
-0.071 |
| 261 |
249293_at |
AT5G41260
|
BSK8, brassinosteroid-signaling kinase 8 |
0.574 |
0.210 |
| 262 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.570 |
0.416 |
| 263 |
248911_at |
AT5G45830
|
ATDOG1, DOG1, DELAY OF GERMINATION 1, GAAS5, germination ability after storage 5, GSQ5, GLUCOSE SENSING QTL 5 |
0.570 |
0.002 |
| 264 |
261580_at |
AT1G01110
|
IQD18, IQ-domain 18 |
0.568 |
-0.055 |
| 265 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
0.564 |
-0.003 |
| 266 |
252100_at |
AT3G51110
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.563 |
-0.112 |
| 267 |
262805_at |
AT1G20900
|
AHL27, AT-hook motif nuclear-localized protein 27, ESC, ESCAROLA, ORE7, ORESARA 7 |
0.561 |
-0.120 |
| 268 |
246562_at |
AT5G15580
|
LNG1, LONGIFOLIA1, TRM2, TON1 Recruiting Motif 2 |
0.561 |
0.034 |
| 269 |
246284_at |
AT4G36780
|
BEH2, BES1/BZR1 homolog 2 |
0.560 |
0.016 |
| 270 |
254797_at |
AT4G13030
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.560 |
0.016 |
| 271 |
259911_at |
AT1G72680
|
ATCAD1, CINNAMYL ALCOHOL DEHYDROGENASE 1, CAD1, cinnamyl-alcohol dehydrogenase |
0.560 |
0.336 |
| 272 |
245276_at |
AT4G16780
|
ATHB-2, homeobox protein 2, ATHB2, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2, HAT4, HB-2, homeobox protein 2 |
0.560 |
-0.080 |
| 273 |
267532_at |
AT2G42040
|
unknown |
0.559 |
0.115 |
| 274 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
0.559 |
0.053 |
| 275 |
262296_at |
AT1G27630
|
CYCT1;3, cyclin T 1;3 |
0.558 |
0.268 |
| 276 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.557 |
-0.007 |
| 277 |
245335_at |
AT4G16160
|
ATOEP16-2, ATOEP16-S |
0.556 |
0.017 |
| 278 |
263680_at |
AT1G26930
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.555 |
0.080 |
| 279 |
253672_at |
AT4G29820
|
ATCFIM-25, ARABIDOPSIS THALIANA HOMOLOG OF CFIM-25, CFIM-25, homolog of CFIM-25 |
0.555 |
0.142 |
| 280 |
259992_at |
AT1G67970
|
AT-HSFA8, HSFA8, heat shock transcription factor A8 |
0.554 |
0.463 |
| 281 |
247793_at |
AT5G58650
|
PSY1, plant peptide containing sulfated tyrosine 1 |
0.554 |
-0.235 |
| 282 |
264525_at |
AT1G10060
|
ATBCAT-1, branched-chain amino acid transaminase 1, BCAT-1, branched-chain amino acid transaminase 1, BCAT1, branched-chain amino acid transferase 1 |
0.554 |
-0.038 |
| 283 |
253331_at |
AT4G33490
|
[Eukaryotic aspartyl protease family protein] |
0.553 |
0.233 |
| 284 |
257122_at |
AT3G20250
|
APUM5, pumilio 5, PUM5, pumilio 5 |
0.552 |
0.044 |
| 285 |
267280_at |
AT2G19450
|
ABX45, AS11, ATDGAT, Arabidopsis thaliana acyl-CoA:diacylglycerol acyltransferase, AtDGAT1, DGAT1, acyl-CoA:diacylglycerol acyltransferase 1, RDS1, TAG1, TRIACYLGLYCEROL BIOSYNTHESIS DEFECT 1 |
0.550 |
0.293 |
| 286 |
264889_at |
AT1G23050
|
hydroxyproline-rich glycoprotein family protein |
0.550 |
-0.189 |
| 287 |
261453_at |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
0.548 |
0.282 |
| 288 |
255255_at |
AT4G05070
|
[Wound-responsive family protein] |
0.548 |
0.078 |
| 289 |
256455_at |
AT1G75190
|
unknown |
0.547 |
0.365 |
| 290 |
247524_at |
AT5G61440
|
ACHT5, atypical CYS HIS rich thioredoxin 5 |
0.547 |
0.048 |
| 291 |
249999_at |
AT5G18640
|
[alpha/beta-Hydrolases superfamily protein] |
0.547 |
0.127 |
| 292 |
247786_at |
AT5G58600
|
PMR5, POWDERY MILDEW RESISTANT 5, TBL44, TRICHOME BIREFRINGENCE-LIKE 44 |
0.547 |
0.317 |
| 293 |
249755_at |
AT5G24580
|
[Heavy metal transport/detoxification superfamily protein ] |
0.546 |
-0.159 |
| 294 |
258527_at |
AT3G06850
|
BCE2, DIN3, DARK INDUCIBLE 3, LTA1 |
0.544 |
0.039 |
| 295 |
261253_at |
AT1G05840
|
[Eukaryotic aspartyl protease family protein] |
0.544 |
0.083 |
| 296 |
250108_at |
AT5G15150
|
ATHB-3, homeobox 3, ATHB3, HAT7, HOMEOBOX FROM ARABIDOPSIS THALIANA 7, HB-3, homeobox 3 |
0.543 |
-0.016 |
| 297 |
251037_at |
AT5G02100
|
ORP3A, OSBP(OXYSTEROL BINDING PROTEIN)-RELATED PROTEIN 3A, UNE18, UNFERTILIZED EMBRYO SAC 18 |
0.543 |
0.142 |
| 298 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
0.542 |
0.060 |
| 299 |
245064_at |
AT2G39725
|
[LYR family of Fe/S cluster biogenesis protein] |
0.541 |
0.167 |
| 300 |
263065_at |
AT2G18170
|
ATMPK7, MAP kinase 7, MPK7, MAP kinase 7 |
0.540 |
0.230 |
| 301 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-1.392 |
-0.935 |
| 302 |
252661_at |
AT3G44450
|
unknown |
-1.385 |
-0.039 |
| 303 |
256096_at |
AT1G13650
|
unknown |
-1.329 |
-0.107 |
| 304 |
257021_at |
AT3G19710
|
BCAT4, branched-chain aminotransferase4 |
-1.316 |
-0.024 |
| 305 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-1.274 |
0.017 |
| 306 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
-1.261 |
-0.191 |
| 307 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
-1.243 |
-0.186 |
| 308 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
-1.190 |
-0.297 |
| 309 |
263829_at |
AT2G40435
|
unknown |
-1.183 |
-0.102 |
| 310 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
-1.179 |
-0.097 |
| 311 |
255088_at |
AT4G09350
|
CRRJ, CHLORORESPIRATORY REDUCTION J, DJC75, DNA J protein C75, NdhT, NADH dehydrogenase-like complex T |
-1.172 |
-0.206 |
| 312 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
-1.171 |
-0.109 |
| 313 |
266326_at |
AT2G46650
|
ATCB5-C, ARABIDOPSIS CYTOCHROME B5 ISOFORM C, B5 #1, CB5-C, cytochrome B5 isoform C, CYTB5-C |
-1.161 |
-0.035 |
| 314 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
-1.151 |
-0.119 |
| 315 |
256603_at |
AT3G28270
|
unknown |
-1.149 |
-0.509 |
| 316 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
-1.144 |
-0.375 |
| 317 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
-1.100 |
-0.035 |
| 318 |
248961_at |
AT5G45650
|
[subtilase family protein] |
-1.099 |
0.014 |
| 319 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-1.098 |
-0.107 |
| 320 |
252363_at |
AT3G48460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-1.097 |
-0.262 |
| 321 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
-1.092 |
-0.373 |
| 322 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
-1.089 |
-0.452 |
| 323 |
257793_at |
AT3G26960
|
[Pollen Ole e 1 allergen and extensin family protein] |
-1.084 |
-0.157 |
| 324 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-1.077 |
-0.202 |
| 325 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
-1.075 |
-0.252 |
| 326 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
-1.074 |
0.096 |
| 327 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-1.066 |
-0.438 |
| 328 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-1.066 |
-0.266 |
| 329 |
252870_at |
AT4G39940
|
AKN2, APS-kinase 2, APK2, ADENOSINE-5'-PHOSPHOSULFATE (APS) KINASE 2 |
-1.050 |
-0.071 |
| 330 |
259348_at |
AT3G03770
|
[Leucine-rich repeat protein kinase family protein] |
-1.041 |
-0.156 |
| 331 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-1.040 |
0.185 |
| 332 |
261913_at |
AT1G65860
|
FMO GS-OX1, flavin-monooxygenase glucosinolate S-oxygenase 1 |
-1.038 |
-0.150 |
| 333 |
249614_at |
AT5G37300
|
WSD1 |
-1.036 |
-0.571 |
| 334 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
-1.025 |
-0.636 |
| 335 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
-1.023 |
-0.296 |
| 336 |
254371_at |
AT4G21760
|
BGLU47, beta-glucosidase 47 |
-1.020 |
-0.343 |
| 337 |
257057_at |
AT3G15310
|
unknown |
-1.015 |
-0.444 |
| 338 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
-1.011 |
-0.083 |
| 339 |
251028_at |
AT5G02230
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-1.010 |
-0.215 |
| 340 |
255786_at |
AT1G19670
|
ATCLH1, chlorophyllase 1, ATHCOR1, CORONATINE-INDUCED PROTEIN 1, CLH1, chlorophyllase 1, CORI1, CORONATINE-INDUCED PROTEIN 1 |
-1.009 |
-0.086 |
| 341 |
250598_at |
AT5G07690
|
ATMYB29, myb domain protein 29, MYB29, myb domain protein 29, PMG2, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 2 |
-1.003 |
-0.093 |
| 342 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
-1.002 |
-0.069 |
| 343 |
265121_at |
AT1G62560
|
FMO GS-OX3, flavin-monooxygenase glucosinolate S-oxygenase 3 |
-0.994 |
-0.086 |
| 344 |
250633_at |
AT5G07460
|
ATMSRA2, ARABIDOPSIS THALIANA METHIONINE SULFOXIDE REDUCTASE 2, PMSR2, peptidemethionine sulfoxide reductase 2 |
-0.994 |
-0.111 |
| 345 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
-0.988 |
-0.438 |
| 346 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
-0.983 |
-0.254 |
| 347 |
254687_at |
AT4G13770
|
CYP83A1, cytochrome P450, family 83, subfamily A, polypeptide 1, REF2, REDUCED EPIDERMAL FLUORESCENCE 2 |
-0.973 |
-0.010 |
| 348 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
-0.970 |
-0.129 |
| 349 |
246021_at |
AT5G21100
|
[Plant L-ascorbate oxidase] |
-0.967 |
-0.230 |
| 350 |
247463_at |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
-0.965 |
-0.379 |
| 351 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
-0.964 |
-0.138 |
| 352 |
260955_at |
AT1G06000
|
[UDP-Glycosyltransferase superfamily protein] |
-0.957 |
-0.228 |
| 353 |
254773_at |
AT4G13410
|
ATCSLA15, CSLA15, CELLULOSE SYNTHASE LIKE A15 |
-0.955 |
-0.148 |
| 354 |
263739_at |
AT2G21320
|
BBX18, B-box domain protein 18 |
-0.952 |
-0.137 |
| 355 |
252073_at |
AT3G51750
|
unknown |
-0.948 |
-0.017 |
| 356 |
265892_at |
AT2G15020
|
unknown |
-0.947 |
-0.314 |
| 357 |
259275_at |
AT3G01060
|
unknown |
-0.946 |
-0.099 |
| 358 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
-0.944 |
0.146 |
| 359 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.943 |
-0.396 |
| 360 |
247214_at |
AT5G64850
|
unknown |
-0.939 |
-0.170 |
| 361 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
-0.937 |
-0.484 |
| 362 |
253496_at |
AT4G31870
|
ATGPX7, GLUTATHIONE PEROXIDASE 7, GPX7, glutathione peroxidase 7 |
-0.930 |
-0.201 |
| 363 |
260745_at |
AT1G78370
|
ATGSTU20, glutathione S-transferase TAU 20, GSTU20, glutathione S-transferase TAU 20 |
-0.922 |
0.013 |
| 364 |
263034_at |
AT1G24020
|
MLP423, MLP-like protein 423 |
-0.921 |
-0.020 |
| 365 |
262883_at |
AT1G64780
|
AMT1;2, ammonium transporter 1;2, ATAMT1;2, ammonium transporter 1;2 |
-0.920 |
-0.433 |
| 366 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
-0.918 |
-0.137 |
| 367 |
247977_at |
AT5G56850
|
unknown |
-0.904 |
-0.137 |
| 368 |
258396_at |
AT3G15460
|
[Ribosomal RNA processing Brix domain protein] |
-0.900 |
0.008 |
| 369 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
-0.899 |
-0.370 |
| 370 |
248867_at |
AT5G46830
|
ATNIG1, NACL-INDUCIBLE GENE 1, NIG1, NACL-inducible gene 1 |
-0.898 |
-0.530 |
| 371 |
253579_at |
AT4G30610
|
BRS1, BRI1 SUPPRESSOR 1, SCPL24, SERINE CARBOXYPEPTIDASE 24 PRECURSOR |
-0.894 |
-0.396 |
| 372 |
252377_at |
AT3G47960
|
AtNPF2.10, GTR1, GLUCOSINOLATE TRANSPORTER-1, NPF2.10, NRT1/ PTR family 2.10 |
-0.892 |
-0.030 |
| 373 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-0.891 |
-0.247 |
| 374 |
255773_at |
AT1G18590
|
ATSOT17, SULFOTRANSFERASE 17, ATST5C, ARABIDOPSIS SULFOTRANSFERASE 5C, SOT17, sulfotransferase 17 |
-0.891 |
-0.080 |
| 375 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.890 |
-0.074 |
| 376 |
249774_at |
AT5G24150
|
SQE5, SQUALENE MONOOXYGENASE 5, SQP1 |
-0.889 |
-0.474 |
| 377 |
255284_at |
AT4G04610
|
APR, APR1, APS reductase 1, ATAPR1, PRH19, PAPS REDUCTASE HOMOLOG 19 |
-0.886 |
-0.020 |
| 378 |
254783_at |
AT4G12830
|
[alpha/beta-Hydrolases superfamily protein] |
-0.881 |
-0.205 |
| 379 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
-0.878 |
-0.191 |
| 380 |
256427_at |
AT3G11090
|
LBD21, LOB domain-containing protein 21 |
-0.876 |
0.001 |
| 381 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.874 |
-0.268 |
| 382 |
264436_at |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
-0.870 |
-0.313 |
| 383 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
-0.869 |
-0.049 |
| 384 |
252414_at |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
-0.866 |
-0.096 |
| 385 |
254862_at |
AT4G12030
|
BASS5, BILE ACID:SODIUM SYMPORTER FAMILY PROTEIN 5, BAT5, bile acid transporter 5 |
-0.856 |
-0.052 |
| 386 |
255298_at |
AT4G04840
|
ATMSRB6, methionine sulfoxide reductase B6, MSRB6, methionine sulfoxide reductase B6 |
-0.855 |
-0.202 |
| 387 |
258370_at |
AT3G14395
|
unknown |
-0.853 |
-0.446 |
| 388 |
261016_at |
AT1G26560
|
BGLU40, beta glucosidase 40 |
-0.846 |
-0.077 |
| 389 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
-0.843 |
-0.253 |
| 390 |
254145_at |
AT4G24700
|
unknown |
-0.837 |
-0.333 |
| 391 |
265939_at |
AT2G19650
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.827 |
-0.251 |
| 392 |
249866_at |
AT5G23010
|
IMS3, 2-ISOPROPYLMALATE SYNTHASE 3, MAM1, methylthioalkylmalate synthase 1 |
-0.826 |
-0.144 |
| 393 |
261150_at |
AT1G19640
|
JMT, jasmonic acid carboxyl methyltransferase |
-0.822 |
-0.378 |
| 394 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
-0.819 |
-0.254 |
| 395 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.818 |
-0.141 |
| 396 |
256304_at |
AT1G69523
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.802 |
-0.634 |
| 397 |
252010_at |
AT3G52740
|
unknown |
-0.800 |
-0.094 |
| 398 |
246516_at |
AT5G15740
|
[O-fucosyltransferase family protein] |
-0.799 |
-0.289 |
| 399 |
264271_at |
AT1G60270
|
BGLU6, beta glucosidase 6 |
-0.798 |
-0.432 |
| 400 |
248246_at |
AT5G53200
|
TRY, TRIPTYCHON |
-0.798 |
-0.330 |
| 401 |
253949_at |
AT4G26780
|
AR192, MGE2, mitochondrial GrpE 2 |
-0.794 |
0.115 |
| 402 |
256673_at |
AT3G52370
|
FLA15, FASCICLIN-like arabinogalactan protein 15 precursor |
-0.788 |
-0.209 |
| 403 |
263918_at |
AT2G36590
|
ATPROT3, PROLINE TRANSPORTER 3, ProT3, proline transporter 3 |
-0.787 |
-0.399 |
| 404 |
266395_at |
AT2G43100
|
ATLEUD1, IPMI2, isopropylmalate isomerase 2 |
-0.781 |
-0.022 |
| 405 |
262826_at |
AT1G13080
|
CYP71B2, cytochrome P450, family 71, subfamily B, polypeptide 2 |
-0.779 |
-0.164 |
| 406 |
247549_at |
AT5G61420
|
AtMYB28, HAG1, HIGH ALIPHATIC GLUCOSINOLATE 1, MYB28, myb domain protein 28, PMG1, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 1 |
-0.774 |
-0.047 |
| 407 |
262526_at |
AT1G17050
|
AtSPS2, SPS2, solanesyl diphosphate synthase 2 |
-0.772 |
-0.240 |
| 408 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
-0.771 |
-0.611 |
| 409 |
252888_at |
AT4G39210
|
APL3 |
-0.770 |
0.091 |
| 410 |
246439_at |
AT5G17600
|
[RING/U-box superfamily protein] |
-0.770 |
-0.179 |
| 411 |
251309_at |
AT3G61220
|
SDR1, short-chain dehydrogenase/reductase 1 |
-0.768 |
-0.112 |
| 412 |
258055_at |
AT3G16250
|
NDF4, NDH-dependent cyclic electron flow 1, PnsB3, Photosynthetic NDH subcomplex B 3 |
-0.766 |
-0.028 |
| 413 |
248075_at |
AT5G55740
|
CRR21, chlororespiratory reduction 21 |
-0.766 |
-0.169 |
| 414 |
255877_at |
AT2G40460
|
[Major facilitator superfamily protein] |
-0.758 |
-0.119 |
| 415 |
257533_at |
AT3G10840
|
[alpha/beta-Hydrolases superfamily protein] |
-0.755 |
-0.195 |
| 416 |
266996_at |
AT2G34490
|
CYP710A2, cytochrome P450, family 710, subfamily A, polypeptide 2 |
-0.754 |
-0.182 |
| 417 |
261907_at |
AT1G65060
|
4CL3, 4-coumarate:CoA ligase 3 |
-0.750 |
-0.333 |
| 418 |
261804_at |
AT1G30530
|
UGT78D1, UDP-glucosyl transferase 78D1 |
-0.748 |
-0.199 |
| 419 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
-0.745 |
0.065 |
| 420 |
247235_at |
AT5G64580
|
EMB3144, EMBRYO DEFECTIVE 3144 |
-0.739 |
0.057 |
| 421 |
251727_at |
AT3G56290
|
unknown |
-0.738 |
-0.216 |
| 422 |
248607_at |
AT5G49480
|
ATCP1, Ca2+-binding protein 1, CP1, Ca2+-binding protein 1 |
-0.736 |
-0.134 |
| 423 |
267010_at |
AT2G39250
|
SNZ, SCHNARCHZAPFEN |
-0.735 |
-0.198 |
| 424 |
247109_at |
AT5G65870
|
ATPSK5, phytosulfokine 5 precursor, PSK5, PSK5, phytosulfokine 5 precursor |
-0.734 |
-0.114 |
| 425 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
-0.733 |
-0.236 |
| 426 |
258589_at |
AT3G04290
|
ATLTL1, LTL1, Li-tolerant lipase 1 |
-0.732 |
-0.061 |
| 427 |
253835_at |
AT4G27820
|
BGLU9, beta glucosidase 9 |
-0.730 |
-0.083 |
| 428 |
263777_at |
AT2G46450
|
ATCNGC12, cyclic nucleotide-gated channel 12, CNGC12, cyclic nucleotide-gated channel 12 |
-0.729 |
-0.052 |
| 429 |
247486_at |
AT5G62140
|
unknown |
-0.728 |
-0.230 |
| 430 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
-0.728 |
-0.210 |
| 431 |
257300_at |
AT3G28080
|
UMAMIT47, Usually multiple acids move in and out Transporters 47 |
-0.727 |
-0.055 |
| 432 |
263841_at |
AT2G36870
|
AtXTH32, XTH32, xyloglucan endotransglucosylase/hydrolase 32 |
-0.727 |
-0.136 |
| 433 |
253922_at |
AT4G26850
|
VTC2, vitamin c defective 2 |
-0.726 |
-0.435 |
| 434 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
-0.725 |
0.042 |
| 435 |
261330_at |
AT1G44900
|
ATMCM2, MCM2, MINICHROMOSOME MAINTENANCE 2 |
-0.722 |
-0.074 |
| 436 |
264839_at |
AT1G03630
|
POR C, protochlorophyllide oxidoreductase C, PORC |
-0.722 |
0.015 |
| 437 |
262811_at |
AT1G11700
|
unknown |
-0.722 |
-0.281 |
| 438 |
245479_at |
AT4G16140
|
[proline-rich family protein] |
-0.719 |
-0.243 |
| 439 |
247055_at |
AT5G66740
|
unknown |
-0.718 |
-0.523 |
| 440 |
263549_at |
AT2G21650
|
ATRL2, ARABIDOPSIS RAD-LIKE 2, MEE3, MATERNAL EFFECT EMBRYO ARREST 3, RSM1, RADIALIS-LIKE SANT/MYB 1 |
-0.716 |
-0.455 |
| 441 |
259925_at |
AT1G75040
|
PR-5, PR5, pathogenesis-related gene 5 |
-0.714 |
0.402 |
| 442 |
266384_at |
AT2G14660
|
unknown |
-0.714 |
-0.123 |
| 443 |
252911_at |
AT4G39510
|
CYP96A12, cytochrome P450, family 96, subfamily A, polypeptide 12 |
-0.713 |
-0.128 |
| 444 |
256168_at |
AT1G51805
|
[Leucine-rich repeat protein kinase family protein] |
-0.713 |
-0.091 |
| 445 |
264052_at |
AT2G22330
|
CYP79B3, cytochrome P450, family 79, subfamily B, polypeptide 3 |
-0.711 |
-0.205 |
| 446 |
257173_at |
AT3G23810
|
ATSAHH2, S-ADENOSYL-L-HOMOCYSTEINE (SAH) HYDROLASE 2, SAHH2, S-adenosyl-l-homocysteine (SAH) hydrolase 2 |
-0.703 |
-0.068 |
| 447 |
258299_at |
AT3G23410
|
ATFAO3, ARABIDOPSIS FATTY ALCOHOL OXIDASE 3, FAO3, fatty alcohol oxidase 3 |
-0.702 |
-0.073 |
| 448 |
262086_at |
AT1G56050
|
[GTP-binding protein-related] |
-0.701 |
0.040 |
| 449 |
253191_at |
AT4G35350
|
XCP1, xylem cysteine peptidase 1 |
-0.695 |
-0.035 |
| 450 |
260343_at |
AT1G69200
|
FLN2, fructokinase-like 2 |
-0.693 |
0.172 |
| 451 |
254683_at |
AT4G13800
|
unknown |
-0.691 |
-0.224 |
| 452 |
260547_at |
AT2G43550
|
[Scorpion toxin-like knottin superfamily protein] |
-0.691 |
0.011 |
| 453 |
260146_at |
AT1G52770
|
[Phototropic-responsive NPH3 family protein] |
-0.691 |
0.243 |
| 454 |
245165_at |
AT2G33180
|
unknown |
-0.690 |
-0.037 |
| 455 |
248625_at |
AT5G48880
|
KAT5, 3-KETO-ACYL-COENZYME A THIOLASE 5, PKT1, PEROXISOMAL-3-KETO-ACYL-COA THIOLASE 1, PKT2, peroxisomal 3-keto-acyl-CoA thiolase 2 |
-0.689 |
-0.100 |
| 456 |
255016_at |
AT4G10120
|
ATSPS4F |
-0.689 |
-0.209 |
| 457 |
258003_at |
AT3G29030
|
ATEXP5, ARABIDOPSIS THALIANA EXPANSIN 5, ATEXPA5, ARABIDOPSIS THALIANA EXPANSIN A5, ATHEXP ALPHA 1.4, EXP5, EXPANSIN 5, EXPA5, expansin A5 |
-0.688 |
-0.067 |
| 458 |
259432_at |
AT1G01520
|
ASG4, ALTERED SEED GERMINATION 4 |
-0.688 |
-0.623 |
| 459 |
258895_at |
AT3G05600
|
[alpha/beta-Hydrolases superfamily protein] |
-0.687 |
-0.164 |
| 460 |
250149_at |
AT5G14700
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.685 |
-0.118 |
| 461 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
-0.684 |
-0.135 |
| 462 |
250006_at |
AT5G18660
|
PCB2, PALE-GREEN AND CHLOROPHYLL B REDUCED 2 |
-0.683 |
-0.030 |
| 463 |
267294_at |
AT2G23670
|
YCF37, homolog of Synechocystis YCF37 |
-0.683 |
-0.028 |
| 464 |
256379_at |
AT1G66840
|
PMI2, plastid movement impaired 2, WEB2, WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 2 |
-0.682 |
-0.243 |
| 465 |
255957_at |
AT1G22160
|
unknown |
-0.677 |
-0.230 |
| 466 |
258675_at |
AT3G08770
|
LTP6, lipid transfer protein 6 |
-0.676 |
0.003 |
| 467 |
246313_at |
AT1G31920
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.675 |
-0.140 |
| 468 |
263115_at |
AT1G03055
|
AtD27, A. thaliana homolog of rice D27, D27, DWARF27 |
-0.675 |
-0.275 |
| 469 |
248891_at |
AT5G46280
|
MCM3, MINICHROMOSOME MAINTENANCE 3 |
-0.674 |
-0.016 |
| 470 |
265305_at |
AT2G20340
|
AAS, aromatic aldehyde synthase, AtAAS |
-0.671 |
-0.169 |
| 471 |
246597_at |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
-0.671 |
-0.321 |
| 472 |
253040_at |
AT4G37800
|
XTH7, xyloglucan endotransglucosylase/hydrolase 7 |
-0.670 |
-0.115 |
| 473 |
266357_at |
AT2G32290
|
BAM6, beta-amylase 6, BMY5, BETA-AMYLASE 5 |
-0.668 |
-0.217 |
| 474 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
-0.664 |
-0.407 |
| 475 |
245982_at |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
-0.662 |
-0.052 |
| 476 |
250985_at |
AT5G02830
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.661 |
-0.184 |
| 477 |
252619_at |
AT3G45210
|
unknown |
-0.658 |
-0.104 |
| 478 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.658 |
-0.360 |
| 479 |
249727_at |
AT5G35490
|
ATMRU1, ARABIDOPSIS MTO 1 RESPONDING UP 1, MRU1, MTO 1 RESPONDING UP 1 |
-0.657 |
-0.075 |
| 480 |
261488_at |
AT1G14345
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.657 |
-0.133 |
| 481 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
-0.655 |
0.030 |
| 482 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
-0.655 |
-0.347 |
| 483 |
255694_at |
AT4G00050
|
UNE10, unfertilized embryo sac 10 |
-0.654 |
-0.393 |
| 484 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
-0.653 |
-0.364 |
| 485 |
255622_at |
AT4G01070
|
GT72B1, UGT72B1, UDP-GLUCOSE-DEPENDENT GLUCOSYLTRANSFERASE 72 B1 |
-0.653 |
-0.081 |
| 486 |
247452_at |
AT5G62430
|
CDF1, cycling DOF factor 1 |
-0.653 |
-0.179 |
| 487 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
-0.653 |
-0.131 |
| 488 |
245876_at |
AT1G26230
|
Cpn60beta4, chaperonin-60beta4 |
-0.652 |
0.005 |
| 489 |
245676_at |
AT1G56670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.652 |
-0.257 |
| 490 |
251658_at |
AT3G57020
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.651 |
-0.178 |
| 491 |
246687_at |
AT5G33370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.650 |
0.056 |
| 492 |
264442_at |
AT1G27480
|
[alpha/beta-Hydrolases superfamily protein] |
-0.650 |
-0.064 |
| 493 |
251418_at |
AT3G60440
|
[Phosphoglycerate mutase family protein] |
-0.649 |
0.146 |
| 494 |
267549_at |
AT2G32640
|
[Lycopene beta/epsilon cyclase protein] |
-0.648 |
-0.120 |
| 495 |
260696_at |
AT1G32520
|
unknown |
-0.648 |
-0.171 |
| 496 |
263674_at |
AT2G04790
|
unknown |
-0.647 |
-0.239 |
| 497 |
256302_at |
AT1G69526
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.647 |
-0.405 |
| 498 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
-0.647 |
-0.194 |
| 499 |
264083_at |
AT2G31230
|
ATERF15, ethylene-responsive element binding factor 15, ERF15, ethylene-responsive element binding factor 15 |
-0.647 |
-0.013 |
| 500 |
253411_at |
AT4G32980
|
ATH1, homeobox gene 1 |
-0.644 |
-0.061 |
| 501 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
-0.644 |
0.017 |
| 502 |
258315_at |
AT3G16175
|
[Thioesterase superfamily protein] |
-0.639 |
-0.338 |
| 503 |
258025_at |
AT3G19480
|
3-PGDH, PGDH3, phosphoglycerate dehydrogenase 3 |
-0.637 |
-0.159 |
| 504 |
255438_at |
AT4G03070
|
AOP, AOP1, AOP1.1 |
-0.637 |
-0.141 |
| 505 |
245501_at |
AT4G15620
|
[Uncharacterised protein family (UPF0497)] |
-0.637 |
-0.090 |
| 506 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
-0.631 |
-0.404 |
| 507 |
266418_at |
AT2G38750
|
ANNAT4, annexin 4, AtANN4 |
-0.627 |
-0.171 |
| 508 |
253534_at |
AT4G31500
|
ATR4, ALTERED TRYPTOPHAN REGULATION 4, CYP83B1, cytochrome P450, family 83, subfamily B, polypeptide 1, RED1, RED ELONGATED 1, RNT1, RUNT 1, SUR2, SUPERROOT 2 |
-0.627 |
-0.043 |
| 509 |
262745_at |
AT1G28600
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.623 |
-0.021 |
| 510 |
265823_at |
AT2G35760
|
[Uncharacterised protein family (UPF0497)] |
-0.622 |
-0.292 |
| 511 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
-0.620 |
-0.028 |
| 512 |
259660_at |
AT1G55260
|
LTPG6, glycosylphosphatidylinositol-anchored lipid protein transfer 6 |
-0.620 |
-0.124 |
| 513 |
253090_at |
AT4G36360
|
BGAL3, beta-galactosidase 3 |
-0.619 |
-0.116 |
| 514 |
263840_at |
AT2G36885
|
unknown |
-0.619 |
-0.077 |
| 515 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.619 |
0.049 |
| 516 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
-0.618 |
-0.111 |
| 517 |
262290_at |
AT1G70985
|
[hydroxyproline-rich glycoprotein family protein] |
-0.617 |
-0.341 |
| 518 |
250960_at |
AT5G02940
|
unknown |
-0.615 |
-0.008 |
| 519 |
257801_at |
AT3G18750
|
ATWNK6, ARABIDOPSIS THALIANA WITH NO K 6, WNK6, with no lysine (K) kinase 6, ZIK5 |
-0.613 |
-0.281 |
| 520 |
254343_at |
AT4G21990
|
APR3, APS reductase 3, ATAPR3, PRH-26, PRH26, PAPS REDUCTASE HOMOLOG 26 |
-0.612 |
0.046 |
| 521 |
255294_at |
AT4G04750
|
[Major facilitator superfamily protein] |
-0.611 |
-0.078 |
| 522 |
246986_at |
AT5G67280
|
RLK, receptor-like kinase |
-0.611 |
-0.041 |
| 523 |
263677_at |
AT1G04520
|
PDLP2, plasmodesmata-located protein 2 |
-0.611 |
-0.244 |
| 524 |
262376_at |
AT1G72970
|
EDA17, embryo sac development arrest 17, HTH, HOTHEAD |
-0.610 |
-0.234 |
| 525 |
258023_at |
AT3G19450
|
ATCAD4, CAD, CAD-C, CAD4, CINNAMYL ALCOHOL DEHYDROGENASE 4 |
-0.609 |
-0.174 |
| 526 |
263410_at |
AT2G04039
|
unknown |
-0.608 |
-0.117 |
| 527 |
257628_at |
AT3G26290
|
CYP71B26, cytochrome P450, family 71, subfamily B, polypeptide 26 |
-0.606 |
-0.383 |
| 528 |
248582_at |
AT5G49910
|
cpHsc70-2, chloroplast heat shock protein 70-2, HSC70-7, HEAT SHOCK PROTEIN 70-7 |
-0.606 |
-0.026 |
| 529 |
246920_at |
AT5G25090
|
AtENODL13, ENODL13, early nodulin-like protein 13 |
-0.605 |
-0.073 |
| 530 |
255460_at |
AT4G02800
|
unknown |
-0.602 |
-0.075 |
| 531 |
265411_at |
AT2G16630
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.602 |
-0.042 |
| 532 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
-0.601 |
0.043 |
| 533 |
247400_at |
AT5G62840
|
[Phosphoglycerate mutase family protein] |
-0.601 |
-0.071 |
| 534 |
256983_at |
AT3G13470
|
Cpn60beta2, chaperonin-60beta2 |
-0.601 |
0.042 |
| 535 |
248372_at |
AT5G51850
|
TRM24, TON1 Recruiting Motif 24 |
-0.600 |
-0.112 |
| 536 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
-0.600 |
-0.311 |
| 537 |
250968_at |
AT5G02890
|
[HXXXD-type acyl-transferase family protein] |
-0.600 |
0.025 |
| 538 |
256529_at |
AT1G33260
|
[Protein kinase superfamily protein] |
-0.597 |
-0.155 |
| 539 |
251586_at |
AT3G58070
|
GIS, GLABROUS INFLORESCENCE STEMS |
-0.595 |
-0.212 |
| 540 |
259671_at |
AT1G52290
|
AtPERK15, proline-rich extensin-like receptor kinase 15, PERK15, proline-rich extensin-like receptor kinase 15 |
-0.595 |
0.078 |
| 541 |
251759_at |
AT3G55630
|
ATDFD, DHFS-FPGS homolog D, DFD, DHFS-FPGS homolog D, FPGS3, folylpolyglutamate synthetase 3 |
-0.591 |
-0.079 |
| 542 |
258033_at |
AT3G21250
|
ABCC8, ATP-binding cassette C8, ATMRP6, ARABIDOPSIS THALIANA MULTIDRUG RESISTANCE-ASSOCIATED PROTEIN 6, MRP6, multidrug resistance-associated protein 6 |
-0.591 |
-0.207 |
| 543 |
248963_at |
AT5G45700
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.588 |
-0.051 |
| 544 |
249303_at |
AT5G41460
|
unknown |
-0.588 |
-0.229 |
| 545 |
249059_at |
AT5G44530
|
[Subtilase family protein] |
-0.587 |
-0.211 |
| 546 |
246468_at |
AT5G17050
|
UGT78D2, UDP-glucosyl transferase 78D2 |
-0.587 |
-0.127 |
| 547 |
254938_at |
AT4G10770
|
ATOPT7, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 7, OPT7, oligopeptide transporter 7 |
-0.586 |
-0.116 |
| 548 |
248924_at |
AT5G45960
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.586 |
-0.171 |
| 549 |
245743_at |
AT1G51080
|
unknown |
-0.585 |
0.003 |
| 550 |
263252_at |
AT2G31380
|
BBX25, B-box domain protein 25, STH, salt tolerance homologue |
-0.585 |
-0.184 |
| 551 |
254572_at |
AT4G19380
|
[Long-chain fatty alcohol dehydrogenase family protein] |
-0.582 |
-0.134 |
| 552 |
264636_at |
AT1G65490
|
unknown |
-0.582 |
-0.090 |
| 553 |
261660_at |
AT1G18370
|
ATNACK1, ARABIDOPSIS NPK1-ACTIVATING KINESIN 1, HIK, HINKEL, NACK1, NPK1-ACTIVATING KINESIN 1 |
-0.581 |
0.033 |
| 554 |
251730_at |
AT3G56330
|
[N2,N2-dimethylguanosine tRNA methyltransferase] |
-0.581 |
0.227 |
| 555 |
263714_at |
AT2G20610
|
ALF1, ABERRANT LATERAL ROOT FORMATION 1, HLS3, HOOKLESS 3, RTY, ROOTY, RTY1, ROOTY 1, SUR1, SUPERROOT 1 |
-0.579 |
-0.039 |
| 556 |
253580_at |
AT4G30400
|
[RING/U-box superfamily protein] |
-0.578 |
-0.095 |
| 557 |
248169_at |
AT5G54610
|
ANK, ankyrin, BDA1, bian da 2 (becoming big in Chinese) |
-0.577 |
0.152 |
| 558 |
247946_at |
AT5G57180
|
CIA2, chloroplast import apparatus 2 |
-0.577 |
0.030 |
| 559 |
262748_at |
AT1G28610
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.577 |
-0.032 |
| 560 |
253088_at |
AT4G36220
|
CYP84A1, CYTOCHROME P450 84A1, FAH1, ferulic acid 5-hydroxylase 1 |
-0.574 |
0.016 |
| 561 |
249046_at |
AT5G44400
|
[FAD-binding Berberine family protein] |
-0.574 |
-0.035 |
| 562 |
251058_at |
AT5G01790
|
unknown |
-0.574 |
-0.163 |
| 563 |
248094_at |
AT5G55220
|
[trigger factor type chaperone family protein] |
-0.572 |
0.002 |
| 564 |
248028_at |
AT5G55620
|
unknown |
-0.571 |
-0.148 |
| 565 |
258167_at |
AT3G21560
|
BRT1, Bright Trichomes 1, UGT84A2, UDP-glucosyl transferase 84A2 |
-0.570 |
-0.037 |
| 566 |
257474_at |
AT1G80850
|
[DNA glycosylase superfamily protein] |
-0.570 |
-0.129 |
| 567 |
258613_at |
AT3G02870
|
VTC4 |
-0.569 |
-0.024 |
| 568 |
250337_at |
AT5G11790
|
NDL2, N-MYC downregulated-like 2 |
-0.569 |
-0.141 |
| 569 |
245965_at |
AT5G19730
|
[Pectin lyase-like superfamily protein] |
-0.569 |
-0.439 |
| 570 |
258979_at |
AT3G09440
|
[Heat shock protein 70 (Hsp 70) family protein] |
-0.566 |
-0.161 |
| 571 |
258409_at |
AT3G17640
|
[Leucine-rich repeat (LRR) family protein] |
-0.566 |
-0.277 |
| 572 |
250095_at |
AT5G17230
|
PSY, PHYTOENE SYNTHASE |
-0.566 |
-0.083 |
| 573 |
265122_at |
AT1G62540
|
FMO GS-OX2, flavin-monooxygenase glucosinolate S-oxygenase 2 |
-0.565 |
-0.111 |
| 574 |
247040_at |
AT5G67150
|
[HXXXD-type acyl-transferase family protein] |
-0.565 |
-0.085 |
| 575 |
258802_at |
AT3G04650
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
-0.564 |
0.043 |
| 576 |
266089_at |
AT2G38010
|
[Neutral/alkaline non-lysosomal ceramidase] |
-0.563 |
-0.018 |
| 577 |
262232_at |
AT1G68600
|
[Aluminium activated malate transporter family protein] |
-0.563 |
-0.129 |
| 578 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
-0.563 |
-0.274 |
| 579 |
245242_at |
AT1G44446
|
ATCAO, ARABIDOPSIS THALIANA CHLOROPHYLL A OXYGENASE, CAO, CHLOROPHYLL A OXYGENASE, CH1, CHLORINA 1 |
-0.562 |
-0.085 |
| 580 |
263386_at |
AT2G40150
|
TBL28, TRICHOME BIREFRINGENCE-LIKE 28 |
-0.561 |
-0.218 |
| 581 |
249138_at |
AT5G43070
|
WPP1, WPP domain protein 1 |
-0.560 |
0.008 |
| 582 |
256015_at |
AT1G19150
|
LHCA2*1, Lhca6, photosystem I light harvesting complex gene 6 |
-0.560 |
-0.059 |
| 583 |
264873_at |
AT1G24100
|
UGT74B1, UDP-glucosyl transferase 74B1 |
-0.560 |
-0.065 |
| 584 |
257314_at |
AT3G26590
|
[MATE efflux family protein] |
-0.559 |
-0.014 |
| 585 |
260601_at |
AT1G55910
|
ZIP11, zinc transporter 11 precursor |
-0.558 |
0.041 |
| 586 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
-0.558 |
-0.017 |
| 587 |
259736_at |
AT1G64390
|
AtGH9C2, glycosyl hydrolase 9C2, GH9C2, glycosyl hydrolase 9C2 |
-0.557 |
-0.031 |
| 588 |
247420_at |
AT5G63100
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.557 |
-0.085 |
| 589 |
265646_at |
AT2G27360
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.557 |
-0.093 |
| 590 |
264692_at |
AT1G70000
|
[myb-like transcription factor family protein] |
-0.555 |
-0.198 |
| 591 |
246998_at |
AT5G67370
|
CGLD27, CONSERVED IN THE GREEN LINEAGE AND DIATOMS 27 |
-0.554 |
-0.218 |
| 592 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
-0.552 |
-0.016 |
| 593 |
261942_at |
AT1G22590
|
AGL87, AGAMOUS-like 87 |
-0.551 |
-0.193 |
| 594 |
247522_at |
AT5G61340
|
unknown |
-0.551 |
-0.229 |
| 595 |
256125_at |
AT1G18250
|
ATLP-1 |
-0.550 |
-0.081 |
| 596 |
250434_at |
AT5G10390
|
H3.1, histone 3.1 |
-0.550 |
0.004 |
| 597 |
247323_at |
AT5G64170
|
LNK1, night light-inducible and clock-regulated 1 |
-0.549 |
-0.132 |
| 598 |
251720_at |
AT3G56160
|
[Sodium Bile acid symporter family] |
-0.549 |
-0.036 |
| 599 |
254185_at |
AT4G23990
|
ATCSLG3, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE-LIKE G3, CSLG3, cellulose synthase like G3 |
-0.549 |
-0.038 |
| 600 |
265182_at |
AT1G23740
|
AOR, alkenal/one oxidoreductase |
-0.548 |
-0.105 |