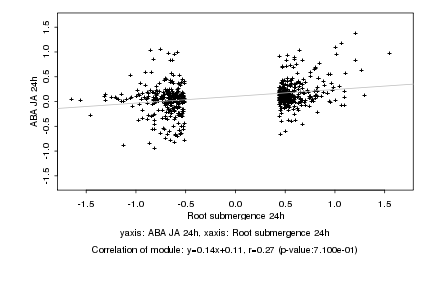
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Root submergence 24h |
Log2 signal ratio
ABA JA 24h |
| 1 |
262325_at |
AT1G64160
|
AtDIR5, DIR5, dirigent protein 5 |
1.544 |
0.981 |
| 2 |
249800_at |
AT5G23660
|
AtSWEET12, MTN3, homolog of Medicago truncatula MTN3, SWEET12 |
1.289 |
0.123 |
| 3 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
1.258 |
0.631 |
| 4 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
1.202 |
0.835 |
| 5 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
1.200 |
1.393 |
| 6 |
258383_at |
AT3G15440
|
unknown |
1.101 |
0.580 |
| 7 |
257163_at |
AT3G24310
|
ATMYB71, MYB DOMAIN PROTEIN 71, MYB305, myb domain protein 305 |
1.095 |
0.083 |
| 8 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
1.092 |
0.219 |
| 9 |
253322_at |
AT4G33980
|
unknown |
1.087 |
-0.068 |
| 10 |
265481_at |
AT2G15960
|
unknown |
1.061 |
1.191 |
| 11 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
1.057 |
-0.062 |
| 12 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
1.036 |
0.321 |
| 13 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
1.014 |
0.966 |
| 14 |
259244_at |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
1.001 |
0.027 |
| 15 |
245528_at |
AT4G15530
|
PPDK, pyruvate orthophosphate dikinase |
0.998 |
1.093 |
| 16 |
263126_at |
AT1G78460
|
[SOUL heme-binding family protein] |
0.977 |
0.286 |
| 17 |
258923_at |
AT3G10450
|
SCPL7, serine carboxypeptidase-like 7 |
0.973 |
0.239 |
| 18 |
267262_at |
AT2G22990
|
SCPL8, SERINE CARBOXYPEPTIDASE-LIKE 8, SNG1, sinapoylglucose 1 |
0.958 |
0.368 |
| 19 |
264774_at |
AT1G22890
|
unknown |
0.955 |
-0.001 |
| 20 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
0.950 |
0.563 |
| 21 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
0.943 |
0.015 |
| 22 |
257832_at |
AT3G26740
|
CCL, CCR-like |
0.932 |
0.551 |
| 23 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
0.921 |
0.167 |
| 24 |
261558_at |
AT1G01770
|
unknown |
0.916 |
0.162 |
| 25 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
0.886 |
0.330 |
| 26 |
264482_at |
AT1G77210
|
AtSTP14, sugar transport protein 14, STP14, sugar transport protein 14 |
0.880 |
0.421 |
| 27 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.878 |
0.187 |
| 28 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.859 |
0.109 |
| 29 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.836 |
0.782 |
| 30 |
258395_at |
AT3G15500
|
ANAC055, NAC domain containing protein 55, ATNAC3, NAC domain containing protein 3, NAC055, NAC domain containing protein 55, NAC3, NAC domain containing protein 3 |
0.828 |
0.471 |
| 31 |
248931_at |
AT5G46040
|
[Major facilitator superfamily protein] |
0.827 |
0.483 |
| 32 |
263198_at |
AT1G53990
|
GLIP3, GDSL-motif lipase 3 |
0.824 |
0.292 |
| 33 |
250868_at |
AT5G03860
|
MLS, malate synthase |
0.820 |
0.106 |
| 34 |
258983_at |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
0.818 |
0.209 |
| 35 |
246389_at |
AT1G77380
|
AAP3, amino acid permease 3, ATAAP3 |
0.816 |
-0.207 |
| 36 |
263295_at |
AT2G14210
|
AGL44, AGAMOUS-like 44, ANR1, ARABIDOPSIS NITRATE REGULATED 1 |
0.810 |
0.226 |
| 37 |
265998_at |
AT2G24270
|
ALDH11A3, aldehyde dehydrogenase 11A3 |
0.807 |
0.136 |
| 38 |
245123_at |
AT2G47450
|
CAO, CHAOS, CPSRP43, CHLOROPLAST SIGNAL RECOGNITION PARTICLE 43 |
0.804 |
0.113 |
| 39 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
0.804 |
0.702 |
| 40 |
259982_at |
AT1G76410
|
ATL8 |
0.800 |
0.680 |
| 41 |
258338_at |
AT3G16150
|
ASPGB1, asparaginase B1 |
0.798 |
0.120 |
| 42 |
262580_at |
AT1G15330
|
[Cystathionine beta-synthase (CBS) protein] |
0.795 |
0.081 |
| 43 |
259316_at |
AT3G01175
|
unknown |
0.795 |
0.130 |
| 44 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
0.789 |
0.664 |
| 45 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
0.785 |
0.203 |
| 46 |
260795_at |
AT1G06225
|
CLE3, CLAVATA3/ESR-RELATED 3 |
0.784 |
0.020 |
| 47 |
247326_at |
AT5G64110
|
[Peroxidase superfamily protein] |
0.769 |
0.050 |
| 48 |
250772_at |
AT5G05420
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
0.758 |
0.016 |
| 49 |
261572_at |
AT1G01170
|
unknown |
0.752 |
0.102 |
| 50 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.751 |
0.595 |
| 51 |
265070_at |
AT1G55510
|
BCDH BETA1, branched-chain alpha-keto acid decarboxylase E1 beta subunit |
0.746 |
0.515 |
| 52 |
248735_at |
AT5G48100
|
ATLAC15, AtTT10, LAC15, LACCASE-LIKE 15, TT10, TRANSPARENT TESTA 10 |
0.745 |
0.083 |
| 53 |
250649_at |
AT5G06690
|
WCRKC1, WCRKC thioredoxin 1 |
0.745 |
0.115 |
| 54 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
0.742 |
0.073 |
| 55 |
250425_at |
AT5G10530
|
LecRK-IX.1, L-type lectin receptor kinase IX.1 |
0.740 |
-0.082 |
| 56 |
248744_at |
AT5G48250
|
BBX8, B-box domain protein 8 |
0.739 |
0.116 |
| 57 |
250940_at |
AT5G03310
|
SAUR44, SMALL AUXIN UPREGULATED RNA 44 |
0.733 |
0.193 |
| 58 |
257615_at |
AT3G26510
|
[Octicosapeptide/Phox/Bem1p family protein] |
0.716 |
0.237 |
| 59 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
0.703 |
-0.066 |
| 60 |
259694_at |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
0.702 |
0.401 |
| 61 |
260688_at |
AT1G17665
|
unknown |
0.696 |
0.022 |
| 62 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
0.690 |
-0.005 |
| 63 |
246951_at |
AT5G04880
|
[expressed protein] |
0.690 |
0.168 |
| 64 |
248138_at |
AT5G54960
|
PDC2, pyruvate decarboxylase-2 |
0.686 |
0.162 |
| 65 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
0.681 |
0.183 |
| 66 |
262322_at |
AT1G27590
|
unknown |
0.677 |
0.166 |
| 67 |
248050_at |
AT5G56100
|
[glycine-rich protein / oleosin] |
0.677 |
0.463 |
| 68 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
0.672 |
0.034 |
| 69 |
261821_at |
AT1G11530
|
ATCXXS1, C-terminal cysteine residue is changed to a serine 1, CXXS1, C-terminal cysteine residue is changed to a serine 1 |
0.671 |
0.273 |
| 70 |
253036_at |
AT4G38340
|
NLP3, NIN-like protein 3 |
0.671 |
-0.445 |
| 71 |
246432_at |
AT5G17490
|
AtRGL3, RGL3, RGA-like protein 3 |
0.666 |
0.347 |
| 72 |
252137_at |
AT3G50980
|
XERO1, dehydrin xero 1 |
0.666 |
0.838 |
| 73 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
0.665 |
0.038 |
| 74 |
255893_at |
AT1G17960
|
[Threonyl-tRNA synthetase] |
0.657 |
-0.067 |
| 75 |
266355_at |
AT2G01400
|
unknown |
0.650 |
0.191 |
| 76 |
263653_at |
AT1G04310
|
ERS2, ethylene response sensor 2 |
0.650 |
0.050 |
| 77 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.649 |
0.081 |
| 78 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
0.649 |
0.374 |
| 79 |
250766_at |
AT5G05550
|
[sequence-specific DNA binding transcription factors] |
0.644 |
0.003 |
| 80 |
263556_at |
AT2G16365
|
[F-box family protein] |
0.644 |
-0.011 |
| 81 |
260741_at |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
0.636 |
1.030 |
| 82 |
252539_at |
AT3G45730
|
unknown |
0.634 |
0.315 |
| 83 |
261842_at |
AT1G16160
|
WAKL5, wall associated kinase-like 5 |
0.633 |
-0.028 |
| 84 |
257922_at |
AT3G23150
|
ETR2, ethylene response 2 |
0.633 |
-0.172 |
| 85 |
256697_at |
AT3G20660
|
AtOCT4, organic cation/carnitine transporter4, OCT4, organic cation/carnitine transporter4 |
0.630 |
0.423 |
| 86 |
262546_at |
AT1G31260
|
ZIP10, zinc transporter 10 precursor |
0.629 |
0.112 |
| 87 |
262635_at |
AT1G06570
|
HPD, 4-hydroxyphenylpyruvate dioxygenase, HPPD, 4-hydroxyphenylpyruvate dioxygenase, PDS1, phytoene desaturation 1 |
0.628 |
0.387 |
| 88 |
256698_at |
AT3G20680
|
unknown |
0.628 |
0.249 |
| 89 |
261613_at |
AT1G49720
|
ABF1, abscisic acid responsive element-binding factor 1, AtABF1 |
0.628 |
0.142 |
| 90 |
260662_at |
AT1G19540
|
[NmrA-like negative transcriptional regulator family protein] |
0.622 |
0.576 |
| 91 |
267493_at |
AT2G30400
|
ATOFP2, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 2, OFP2, ovate family protein 2 |
0.620 |
0.090 |
| 92 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
0.617 |
-0.140 |
| 93 |
258527_at |
AT3G06850
|
BCE2, DIN3, DARK INDUCIBLE 3, LTA1 |
0.605 |
0.373 |
| 94 |
253666_at |
AT4G30270
|
MERI-5, MERISTEM 5, MERI5B, meristem-5, SEN4, SENESCENCE 4, XTH24, xyloglucan endotransglucosylase/hydrolase 24 |
0.604 |
0.750 |
| 95 |
260529_at |
AT2G47400
|
CP12, CP12 DOMAIN-CONTAINING PROTEIN 1, CP12-1, CP12 domain-containing protein 1 |
0.603 |
-0.006 |
| 96 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
0.600 |
0.288 |
| 97 |
264551_at |
AT1G09460
|
[Carbohydrate-binding X8 domain superfamily protein] |
0.600 |
0.083 |
| 98 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.599 |
0.347 |
| 99 |
263836_at |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
0.596 |
-0.376 |
| 100 |
266925_at |
AT2G45740
|
PEX11D, peroxin 11D |
0.593 |
0.209 |
| 101 |
262730_at |
AT1G16390
|
ATOCT3, organic cation/carnitine transporter 3, OCT3, organic cation/carnitine transporter 3 |
0.591 |
0.223 |
| 102 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
0.591 |
-0.093 |
| 103 |
254384_at |
AT4G21870
|
[HSP20-like chaperones superfamily protein] |
0.591 |
0.042 |
| 104 |
261211_at |
AT1G12780
|
ATUGE1, A. THALIANA UDP-GLC 4-EPIMERASE 1, UGE1, UDP-D-glucose/UDP-D-galactose 4-epimerase 1 |
0.590 |
0.361 |
| 105 |
249234_at |
AT5G42200
|
[RING/U-box superfamily protein] |
0.590 |
0.294 |
| 106 |
248975_at |
AT5G45040
|
CYTC6A, cytochrome c6A |
0.589 |
0.116 |
| 107 |
254001_at |
AT4G26260
|
MIOX4, myo-inositol oxygenase 4 |
0.587 |
0.866 |
| 108 |
252746_at |
AT3G43190
|
ATSUS4, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 4, SUS4, sucrose synthase 4 |
0.585 |
0.890 |
| 109 |
254652_at |
AT4G18170
|
ATWRKY28, WRKY28, WRKY DNA-binding protein 28 |
0.584 |
-0.253 |
| 110 |
256499_at |
AT1G36640
|
unknown |
0.583 |
0.121 |
| 111 |
245838_at |
AT1G58410
|
[Disease resistance protein (CC-NBS-LRR class) family] |
0.582 |
0.001 |
| 112 |
245543_at |
AT4G15260
|
[UDP-Glycosyltransferase superfamily protein] |
0.580 |
0.111 |
| 113 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
0.578 |
0.055 |
| 114 |
255181_at |
AT4G08110
|
[CACTA-like transposase family (Ptta/En/Spm), has a 4.2e-66 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
0.576 |
-0.151 |
| 115 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
0.575 |
0.699 |
| 116 |
262629_at |
AT1G06460
|
ACD31.2, ALPHA-CRYSTALLIN DOMAIN 31.2, ACD32.1, alpha-crystallin domain 32.1 |
0.573 |
0.221 |
| 117 |
260115_at |
AT1G33870
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.571 |
0.020 |
| 118 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.570 |
0.129 |
| 119 |
259441_at |
AT1G02300
|
[Cysteine proteinases superfamily protein] |
0.568 |
0.141 |
| 120 |
247136_at |
AT5G66170
|
STR18, sulfurtransferase 18 |
0.566 |
0.469 |
| 121 |
248347_at |
AT5G52250
|
EFO1, EARLY FLOWERING BY OVEREXPRESSION 1, RUP1, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 |
0.564 |
0.198 |
| 122 |
250773_at |
AT5G05430
|
[RNA-binding protein] |
0.562 |
-0.029 |
| 123 |
251100_at |
AT5G01670
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.562 |
0.045 |
| 124 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.557 |
0.383 |
| 125 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
0.557 |
-0.393 |
| 126 |
254139_at |
AT4G24600
|
unknown |
0.555 |
0.093 |
| 127 |
249869_at |
AT5G23050
|
AAE17, acyl-activating enzyme 17 |
0.555 |
0.441 |
| 128 |
263350_at |
AT2G13360
|
AGT, alanine:glyoxylate aminotransferase, AGT1, ALANINE:GLYOXYLATE AMINOTRANSFERASE 1, SGAT, L-serine:glyoxylate aminotransferase |
0.553 |
0.427 |
| 129 |
260411_at |
AT1G69890
|
unknown |
0.553 |
0.330 |
| 130 |
264525_at |
AT1G10060
|
ATBCAT-1, branched-chain amino acid transaminase 1, BCAT-1, branched-chain amino acid transaminase 1, BCAT1, branched-chain amino acid transferase 1 |
0.552 |
0.080 |
| 131 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
0.551 |
0.745 |
| 132 |
264905_at |
AT2G17430
|
ATMLO7, MLO7, MILDEW RESISTANCE LOCUS O 7, NTA, NORTIA |
0.551 |
0.140 |
| 133 |
260451_at |
AT1G72360
|
AtERF73, ERF73, ethylene response factor 73, HRE1, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 1 |
0.550 |
0.323 |
| 134 |
248564_at |
AT5G49700
|
AHL17, AT-hook motif nuclear localized protein 17 |
0.550 |
0.349 |
| 135 |
258281_at |
AT3G26900
|
ATSKL1, Arabidopsis thaliana shikimate kinase-like 1, SKL1, shikimate kinase-like 1 |
0.548 |
0.159 |
| 136 |
252387_at |
AT3G47800
|
[Galactose mutarotase-like superfamily protein] |
0.545 |
0.304 |
| 137 |
263593_at |
AT2G01860
|
EMB975, EMBRYO DEFECTIVE 975 |
0.544 |
0.028 |
| 138 |
258935_at |
AT3G10120
|
unknown |
0.544 |
0.020 |
| 139 |
246703_at |
AT5G28080
|
WNK9 |
0.541 |
0.045 |
| 140 |
261294_at |
AT1G48430
|
[Dihydroxyacetone kinase] |
0.540 |
0.219 |
| 141 |
265992_at |
AT2G24130
|
[Leucine-rich receptor-like protein kinase family protein] |
0.540 |
0.176 |
| 142 |
254409_at |
AT4G21400
|
CRK28, cysteine-rich RLK (RECEPTOR-like protein kinase) 28 |
0.540 |
-0.102 |
| 143 |
249066_at |
AT5G43920
|
[transducin family protein / WD-40 repeat family protein] |
0.538 |
0.002 |
| 144 |
261205_at |
AT1G12790
|
unknown |
0.537 |
0.117 |
| 145 |
251356_at |
AT3G61060
|
AtPP2-A13, phloem protein 2-A13, PP2-A13, phloem protein 2-A13 |
0.536 |
0.334 |
| 146 |
266016_at |
AT2G18670
|
[RING/U-box superfamily protein] |
0.536 |
0.221 |
| 147 |
261100_at |
AT1G63020
|
NRPD1, NRPD1A, nuclear RNA polymerase D1A, POL IVA, NUCLEAR RNA POLYMERASE D 1A, SDE4, SMD2, SILENCING MOVEMENT DEFICIENT 2 |
0.533 |
0.199 |
| 148 |
262162_at |
AT1G78020
|
unknown |
0.533 |
-0.063 |
| 149 |
245925_at |
AT5G28770
|
AtbZIP63, Arabidopsis thaliana basic leucine zipper 63, BZO2H3 |
0.532 |
-0.076 |
| 150 |
267054_at |
AT2G38370
|
unknown |
0.532 |
-0.173 |
| 151 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
0.532 |
0.096 |
| 152 |
248895_at |
AT5G46330
|
FLS2, FLAGELLIN-SENSITIVE 2 |
0.531 |
-0.083 |
| 153 |
252924_at |
AT4G39070
|
BBX20, B-box domain protein 20, BZS1, BZS1 |
0.529 |
0.142 |
| 154 |
250752_at |
AT5G05690
|
CBB3, CABBAGE 3, CPD, CONSTITUTIVE PHOTOMORPHOGENIC DWARF, CYP90, CYP90A, CYP90A1, CYTOCHROME P450 90A1, DWF3, DWARF 3 |
0.527 |
-0.025 |
| 155 |
260522_x_at |
AT2G41730
|
unknown |
0.526 |
-0.372 |
| 156 |
249389_at |
AT5G40100
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.524 |
0.120 |
| 157 |
249824_at |
AT5G23380
|
unknown |
0.523 |
0.166 |
| 158 |
253829_at |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
0.523 |
0.337 |
| 159 |
251884_at |
AT3G54150
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.522 |
0.039 |
| 160 |
252570_at |
AT3G45300
|
ATIVD, IVD, isovaleryl-CoA-dehydrogenase, IVDH, ISOVALERYL-COA-DEHYDROGENASE |
0.522 |
0.413 |
| 161 |
244903_at |
ATMG00660
|
ORF149 |
0.522 |
-0.060 |
| 162 |
264561_at |
AT1G55810
|
UKL3, uridine kinase-like 3 |
0.521 |
0.262 |
| 163 |
265902_at |
AT2G25590
|
[Plant Tudor-like protein] |
0.520 |
0.026 |
| 164 |
263118_at |
AT1G03090
|
MCCA |
0.519 |
0.420 |
| 165 |
264082_at |
AT2G28570
|
unknown |
0.518 |
0.055 |
| 166 |
261618_at |
AT1G33110
|
[MATE efflux family protein] |
0.517 |
0.243 |
| 167 |
247525_at |
AT5G61380
|
APRR1, AtTOC1, TIMING OF CAB EXPRESSION 1, PRR1, PSEUDO-RESPONSE REGULATOR 1, TOC1, TIMING OF CAB EXPRESSION 1 |
0.517 |
-0.019 |
| 168 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.516 |
0.946 |
| 169 |
246251_at |
AT4G37220
|
[Cold acclimation protein WCOR413 family] |
0.514 |
0.466 |
| 170 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
0.513 |
0.158 |
| 171 |
254660_at |
AT4G18250
|
[receptor serine/threonine kinase, putative] |
0.513 |
-0.118 |
| 172 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.512 |
0.232 |
| 173 |
263237_at |
AT1G10610
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.512 |
0.063 |
| 174 |
248395_at |
AT5G52120
|
AtPP2-A14, phloem protein 2-A14, PP2-A14, phloem protein 2-A14 |
0.511 |
0.089 |
| 175 |
253776_at |
AT4G28390
|
AAC3, ADP/ATP carrier 3, ATAAC3 |
0.510 |
0.197 |
| 176 |
264700_at |
AT1G70100
|
unknown |
0.510 |
0.129 |
| 177 |
262502_at |
AT1G21600
|
PTAC6, plastid transcriptionally active 6 |
0.509 |
0.008 |
| 178 |
263909_at |
AT2G36490
|
AtROS1, DML1, demeter-like 1, ROS1, REPRESSOR OF SILENCING1 |
0.508 |
0.094 |
| 179 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
0.505 |
0.838 |
| 180 |
260205_at |
AT1G70700
|
JAZ9, JASMONATE-ZIM-DOMAIN PROTEIN 9, TIFY7 |
0.504 |
0.713 |
| 181 |
263196_at |
AT1G36070
|
[Transducin/WD40 repeat-like superfamily protein] |
0.502 |
0.083 |
| 182 |
256340_at |
AT1G72070
|
[Chaperone DnaJ-domain superfamily protein] |
0.499 |
-0.069 |
| 183 |
249393_at |
AT5G40170
|
AtRLP54, receptor like protein 54, RLP54, receptor like protein 54 |
0.498 |
0.022 |
| 184 |
255148_at |
AT4G08470
|
MAPKKK10, MEKK3, MAPK/ERK kinase kinase 3 |
0.498 |
-0.016 |
| 185 |
257373_at |
AT2G43140
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.497 |
-0.605 |
| 186 |
261588_at |
AT1G01670
|
[RING/U-box superfamily protein] |
0.497 |
0.046 |
| 187 |
247532_at |
AT5G61560
|
[U-box domain-containing protein kinase family protein] |
0.496 |
-0.047 |
| 188 |
249929_at |
AT5G22340
|
unknown |
0.496 |
0.039 |
| 189 |
258100_at |
AT3G23550
|
[MATE efflux family protein] |
0.496 |
0.214 |
| 190 |
254787_at |
AT4G12690
|
unknown |
0.495 |
0.349 |
| 191 |
260272_at |
AT1G80570
|
[RNI-like superfamily protein] |
0.495 |
0.234 |
| 192 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
0.494 |
0.262 |
| 193 |
253160_at |
AT4G35760
|
LTO1, Lumen Thiol Oxidoreductase 1 |
0.494 |
0.119 |
| 194 |
264124_at |
AT1G79360
|
ATOCT2, organic cation/carnitine transporter 2, OCT2, ORGANIC CATION TRANSPORTER 2, OCT2, organic cation/carnitine transporter 2 |
0.493 |
-0.045 |
| 195 |
251082_at |
AT5G01530
|
LHCB4.1, light harvesting complex photosystem II |
0.492 |
0.117 |
| 196 |
249750_at |
AT5G24570
|
unknown |
0.491 |
0.295 |
| 197 |
247617_at |
AT5G60270
|
LecRK-I.7, L-type lectin receptor kinase I.7 |
0.490 |
-0.128 |
| 198 |
248224_at |
AT5G53490
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.489 |
0.137 |
| 199 |
264843_at |
AT1G03400
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.489 |
0.229 |
| 200 |
256965_at |
AT3G13450
|
DIN4, DARK INDUCIBLE 4 |
0.488 |
0.333 |
| 201 |
248193_at |
AT5G54080
|
AtHGO, HGO, homogentisate 1,2-dioxygenase |
0.488 |
0.442 |
| 202 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
0.487 |
-0.102 |
| 203 |
264405_at |
AT1G10210
|
ATMPK1, mitogen-activated protein kinase 1, MPK1, mitogen-activated protein kinase 1 |
0.487 |
0.163 |
| 204 |
264929_at |
AT1G60730
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.487 |
0.282 |
| 205 |
262643_at |
AT1G62770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.484 |
-0.010 |
| 206 |
253722_at |
AT4G29190
|
AtC3H49, AtOZF2, Arabidopsis thaliana oxidation-related zinc finger 2, AtTZF3, OZF2, oxidation-related zinc finger 2, TZF3, tandem zinc finger 3 |
0.483 |
0.201 |
| 207 |
266224_at |
AT2G28800
|
ALB3, ALBINO 3 |
0.482 |
0.169 |
| 208 |
258727_at |
AT3G11930
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.482 |
0.418 |
| 209 |
262452_at |
AT1G11210
|
unknown |
0.481 |
0.293 |
| 210 |
255540_at |
AT4G01800
|
AGY1, Albino or Glassy Yellow 1, AtcpSecA, Arabidopsis thaliana chloroplast SecA, SECA1 |
0.481 |
0.059 |
| 211 |
249415_at |
AT5G39660
|
CDF2, cycling DOF factor 2 |
0.478 |
0.148 |
| 212 |
261542_at |
AT1G63560
|
[Receptor-like protein kinase-related family protein] |
0.476 |
0.062 |
| 213 |
248029_at |
AT5G55700
|
BAM4, beta-amylase 4, BMY6, BETA-AMYLASE 6 |
0.476 |
0.180 |
| 214 |
261518_at |
AT1G71695
|
[Peroxidase superfamily protein] |
0.476 |
0.358 |
| 215 |
255452_at |
AT4G02880
|
unknown |
0.476 |
0.108 |
| 216 |
265189_at |
AT1G23840
|
unknown |
0.474 |
0.229 |
| 217 |
246493_at |
AT5G16180
|
ATCRS1, ARABIDOPSIS ORTHOLOG OF MAIZE CHLOROPLAST SPLICING FACTOR CRS1, CRS1, ortholog of maize chloroplast splicing factor CRS1 |
0.474 |
-0.007 |
| 218 |
248148_at |
AT5G54930
|
[AT hook motif-containing protein] |
0.473 |
-0.016 |
| 219 |
251022_at |
AT5G02150
|
Fes1C, Fes1C |
0.473 |
0.138 |
| 220 |
250913_at |
AT5G03770
|
AtKdtA, KDTA, KDO transferase A |
0.470 |
0.088 |
| 221 |
264580_at |
AT1G05340
|
unknown |
0.470 |
0.435 |
| 222 |
264236_at |
AT1G54680
|
unknown |
0.470 |
0.034 |
| 223 |
260966_at |
AT1G12220
|
RPS5, RESISTANT TO P. SYRINGAE 5 |
0.470 |
0.108 |
| 224 |
246104_at |
AT5G28650
|
ATWRKY74, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 74, WRKY74, WRKY DNA-binding protein 74 |
0.469 |
0.019 |
| 225 |
260837_at |
AT1G43670
|
AtcFBP, Arabidopsis thaliana cytosolic fructose-1,6-bisphosphatase, FBP, fructose-1,6-bisphosphatase, FINS1, FRUCTOSE INSENSITIVE 1 |
0.468 |
0.205 |
| 226 |
246875_at |
AT5G26130
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.467 |
0.083 |
| 227 |
265149_at |
AT1G51400
|
[Photosystem II 5 kD protein] |
0.467 |
0.140 |
| 228 |
260657_at |
AT1G19390
|
[Wall-associated kinase family protein] |
0.467 |
0.195 |
| 229 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
0.466 |
0.710 |
| 230 |
254197_at |
AT4G24040
|
ATTRE1, TRE1, trehalase 1 |
0.466 |
0.205 |
| 231 |
260760_at |
AT1G49170
|
unknown |
0.465 |
0.157 |
| 232 |
250152_at |
AT5G15120
|
unknown |
0.464 |
0.058 |
| 233 |
264836_at |
AT1G03610
|
unknown |
0.463 |
0.162 |
| 234 |
258434_at |
AT3G16770
|
ATEBP, ethylene-responsive element binding protein, EBP, ethylene-responsive element binding protein, ERF72, ETHYLENE RESPONSE FACTOR 72, RAP2.3, RELATED TO AP2 3 |
0.463 |
0.299 |
| 235 |
259058_at |
AT3G03470
|
CYP89A9, cytochrome P450, family 87, subfamily A, polypeptide 9 |
0.463 |
0.698 |
| 236 |
247079_at |
AT5G66055
|
AKRP, ankyrin repeat protein, EMB16, EMBRYO DEFECTIVE 16, EMB2036, EMBRYO DEFECTIVE 2036 |
0.462 |
-0.010 |
| 237 |
261658_at |
AT1G50040
|
unknown |
0.462 |
0.047 |
| 238 |
250248_at |
AT5G13740
|
ZIF1, zinc induced facilitator 1 |
0.462 |
-0.003 |
| 239 |
266839_at |
AT2G25930
|
ELF3, EARLY FLOWERING 3, PYK20 |
0.461 |
0.029 |
| 240 |
260498_at |
AT2G41710
|
[Integrase-type DNA-binding superfamily protein] |
0.460 |
0.045 |
| 241 |
260974_at |
AT1G53440
|
[Leucine-rich repeat transmembrane protein kinase] |
0.460 |
-0.395 |
| 242 |
254032_at |
AT4G25940
|
[ENTH/ANTH/VHS superfamily protein] |
0.460 |
0.066 |
| 243 |
251066_at |
AT5G01880
|
DAFL2, DAF-Like gene 2 |
0.458 |
0.394 |
| 244 |
263490_at |
AT2G42620
|
MAX2, MORE AXILLARY BRANCHES 2, ORE9, ORESARA 9, PPS, PLEIOTROPIC PHOTOSIGNALING |
0.458 |
0.377 |
| 245 |
264199_at |
AT1G22700
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.457 |
0.104 |
| 246 |
263194_at |
AT1G36060
|
[Integrase-type DNA-binding superfamily protein] |
0.457 |
-0.230 |
| 247 |
263708_at |
AT1G09320
|
[agenet domain-containing protein] |
0.456 |
0.023 |
| 248 |
259008_at |
AT3G09390
|
ATMT-1, ARABIDOPSIS THALIANA METALLOTHIONEIN-1, ATMT-K, ARABIDOPSIS THALIANA METALLOTHIONEIN-K, MT2A, metallothionein 2A |
0.455 |
0.029 |
| 249 |
266695_at |
AT2G19810
|
AtOZF1, AtTZF2, OZF1, Oxidation-related Zinc Finger 1, TZF2, tandem zinc finger 2 |
0.455 |
0.273 |
| 250 |
259685_at |
AT1G63090
|
AtPP2-A11, phloem protein 2-A11, PP2-A11, phloem protein 2-A11 |
0.455 |
0.077 |
| 251 |
266917_at |
AT2G45830
|
DTA2, downstream target of AGL15 2 |
0.455 |
0.164 |
| 252 |
251719_at |
AT3G56140
|
unknown |
0.455 |
0.199 |
| 253 |
257698_at |
AT3G12730
|
[Homeodomain-like superfamily protein] |
0.455 |
0.042 |
| 254 |
249485_at |
AT5G39020
|
[Malectin/receptor-like protein kinase family protein] |
0.454 |
-0.137 |
| 255 |
257990_at |
AT3G19860
|
bHLH121, basic Helix-Loop-Helix 121 |
0.453 |
0.142 |
| 256 |
256530_at |
AT1G33290
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.453 |
0.137 |
| 257 |
250776_at |
AT5G05320
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
0.452 |
0.135 |
| 258 |
265330_at |
AT2G18440
|
GUT15, GENE WITH UNSTABLE TRANSCRIPT 15 |
0.451 |
-0.055 |
| 259 |
260799_at |
AT1G78270
|
AtUGT85A4, UDP-glucosyl transferase 85A4, UGT85A4, UDP-glucosyl transferase 85A4 |
0.451 |
-0.001 |
| 260 |
245228_at |
AT3G29810
|
COBL2, COBRA-like protein 2 precursor |
0.451 |
0.191 |
| 261 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
0.450 |
-0.162 |
| 262 |
250414_at |
AT5G11180
|
ATGLR2.6, ARABIDOPSIS THALIANA GLUTAMATE RECEPTOR 2.6, GLR2.6, glutamate receptor 2.6 |
0.449 |
0.151 |
| 263 |
246028_at |
AT5G21170
|
5'-AMP-activated protein kinase beta-2 subunit protein |
0.449 |
0.260 |
| 264 |
264586_at |
AT1G05160
|
ATKAO1, CYP88A3, cytochrome P450, family 88, subfamily A, polypeptide 3, KAO1, ENT-KAURENOIC ACID OXYDASE 1 |
0.449 |
0.087 |
| 265 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
0.449 |
-0.658 |
| 266 |
262505_at |
AT1G21680
|
[DPP6 N-terminal domain-like protein] |
0.448 |
0.279 |
| 267 |
251420_at |
AT3G60490
|
[Integrase-type DNA-binding superfamily protein] |
0.448 |
0.026 |
| 268 |
265535_at |
AT2G15900
|
[Phox-associated domain] |
0.448 |
0.113 |
| 269 |
246922_at |
AT5G25110
|
CIPK25, CBL-interacting protein kinase 25, SnRK3.25, SNF1-RELATED PROTEIN KINASE 3.25 |
0.448 |
0.088 |
| 270 |
257159_at |
AT3G24400
|
AtPERK2, proline-rich extensin-like receptor kinase 2, PERK2, proline-rich extensin-like receptor kinase 2 |
0.447 |
-0.001 |
| 271 |
252990_at |
AT4G38440
|
IYO, MINIYO |
0.447 |
-0.095 |
| 272 |
254545_at |
AT4G19830
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
0.447 |
0.300 |
| 273 |
260160_at |
AT1G79880
|
AtLa2, La2, La protein 2 |
0.447 |
0.069 |
| 274 |
257772_at |
AT3G23080
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.445 |
0.201 |
| 275 |
247540_at |
AT5G61590
|
[Integrase-type DNA-binding superfamily protein] |
0.445 |
0.244 |
| 276 |
260774_at |
AT1G78290
|
SNRK2-8, SNF1-RELATED PROTEIN KINASE 2-8, SNRK2.8, SNF1-RELATED PROTEIN KINASE 2.8, SRK2C, SNF1-RELATED PROTEIN KINASE 2C |
0.445 |
0.000 |
| 277 |
262189_at |
AT1G78010
|
[tRNA modification GTPase, putative] |
0.444 |
0.010 |
| 278 |
245418_at |
AT4G17370
|
[Oxidoreductase family protein] |
0.444 |
0.164 |
| 279 |
260465_at |
AT1G10910
|
EMB3103, EMBRYO DEFECTIVE 3103 |
0.444 |
-0.023 |
| 280 |
251133_at |
AT5G01240
|
LAX1, like AUXIN RESISTANT 1 |
0.444 |
0.194 |
| 281 |
254105_at |
AT4G25080
|
CHLM, magnesium-protoporphyrin IX methyltransferase |
0.440 |
0.126 |
| 282 |
249839_at |
AT5G23405
|
[HMG-box (high mobility group) DNA-binding family protein] |
0.440 |
0.066 |
| 283 |
262884_at |
AT1G64720
|
CP5 |
0.440 |
0.148 |
| 284 |
253624_at |
AT4G30580
|
ATS2, EMB1995, EMBRYO DEFECTIVE 1995, LPAT1, lysophosphatidic acid acyltransferase 1 |
0.440 |
0.022 |
| 285 |
253887_at |
AT4G27730
|
ATOPT6, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 6, OPT6, oligopeptide transporter 1 |
0.439 |
0.033 |
| 286 |
248901_at |
AT5G46410
|
SSP4, SCP1-like small phosphatase 4 |
0.439 |
0.227 |
| 287 |
260876_at |
AT1G21460
|
AtSWEET1, SWEET1 |
0.439 |
0.210 |
| 288 |
256071_at |
AT1G13640
|
[Phosphatidylinositol 3- and 4-kinase family protein] |
0.439 |
-0.050 |
| 289 |
252530_at |
AT3G46500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.439 |
0.050 |
| 290 |
266018_at |
AT2G18710
|
SCY1, SECY homolog 1 |
0.439 |
0.088 |
| 291 |
246284_at |
AT4G36780
|
BEH2, BES1/BZR1 homolog 2 |
0.439 |
0.200 |
| 292 |
247004_at |
AT5G67570
|
DG1, DELAYED GREENING 1, EMB1408, embryo defective 1408, EMB246, EMBRYO DEFECTIVE 246 |
0.438 |
0.040 |
| 293 |
253279_at |
AT4G34030
|
MCCB, 3-methylcrotonyl-CoA carboxylase |
0.436 |
0.200 |
| 294 |
249101_at |
AT5G43580
|
UPI, UNUSUAL SERINE PROTEASE INHIBITOR |
0.436 |
0.914 |
| 295 |
252240_at |
AT3G50010
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.436 |
-0.302 |
| 296 |
262626_at |
AT1G06430
|
FTSH8, FTSH protease 8 |
0.436 |
0.285 |
| 297 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
0.436 |
0.284 |
| 298 |
254206_at |
AT4G24180
|
ATTLP1, TLP1, THAUMATIN-LIKE PROTEIN 1 |
0.436 |
0.099 |
| 299 |
256394_at |
AT3G06290
|
AtSAC3B, SAC3B, yeast Sac3 homolog B |
0.435 |
-0.047 |
| 300 |
266984_at |
AT2G39570
|
ACR9, ACT domain repeats 9 |
0.435 |
0.141 |
| 301 |
253502_at |
AT4G31940
|
CYP82C4, cytochrome P450, family 82, subfamily C, polypeptide 4 |
-1.655 |
0.048 |
| 302 |
254550_at |
AT4G19690
|
ATIRT1, ARABIDOPSIS IRON-REGULATED TRANSPORTER 1, IRT1, iron-regulated transporter 1 |
-1.560 |
0.033 |
| 303 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
-1.462 |
-0.273 |
| 304 |
254907_at |
AT4G11190
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-1.317 |
0.114 |
| 305 |
260745_at |
AT1G78370
|
ATGSTU20, glutathione S-transferase TAU 20, GSTU20, glutathione S-transferase TAU 20 |
-1.313 |
0.149 |
| 306 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
-1.307 |
0.025 |
| 307 |
253040_at |
AT4G37800
|
XTH7, xyloglucan endotransglucosylase/hydrolase 7 |
-1.254 |
0.083 |
| 308 |
257135_at |
AT3G12900
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-1.214 |
0.093 |
| 309 |
254056_at |
AT4G25250
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-1.200 |
0.072 |
| 310 |
262373_at |
AT1G73120
|
unknown |
-1.178 |
0.060 |
| 311 |
253684_at |
AT4G29690
|
[Alkaline-phosphatase-like family protein] |
-1.150 |
0.017 |
| 312 |
250142_at |
AT5G14650
|
[Pectin lyase-like superfamily protein] |
-1.146 |
0.153 |
| 313 |
259388_at |
AT1G13420
|
ATST4B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 4B, ST4B, sulfotransferase 4B |
-1.135 |
0.004 |
| 314 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
-1.129 |
-0.868 |
| 315 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
-1.104 |
0.060 |
| 316 |
260067_at |
AT1G73780
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-1.064 |
0.069 |
| 317 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
-1.058 |
0.537 |
| 318 |
246229_at |
AT4G37160
|
sks15, SKU5 similar 15 |
-1.045 |
0.100 |
| 319 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
-1.036 |
-0.091 |
| 320 |
247483_at |
AT5G62420
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.997 |
0.103 |
| 321 |
248728_at |
AT5G48000
|
CYP708 A2, CYTOCHROME P450, FAMILY 708, SUBFAMILY A, POLYPEPTIDE 2, CYP708A2, cytochrome P450, family 708, subfamily A, polypeptide 2, THAH, THALIANOL HYDROXYLASE, THAH1, THALIANOL HYDROXYLASE 1 |
-0.987 |
0.236 |
| 322 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.983 |
-0.374 |
| 323 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
-0.982 |
0.148 |
| 324 |
267222_at |
AT2G43880
|
[Pectin lyase-like superfamily protein] |
-0.973 |
-0.054 |
| 325 |
254909_at |
AT4G11210
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.969 |
0.139 |
| 326 |
267101_at |
AT2G41480
|
AtPRX25, PRX25, peroxidase 25 |
-0.957 |
0.372 |
| 327 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
-0.951 |
0.042 |
| 328 |
247070_at |
AT5G66815
|
unknown |
-0.947 |
0.046 |
| 329 |
249599_at |
AT5G37990
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.941 |
0.168 |
| 330 |
250661_at |
AT5G07030
|
[Eukaryotic aspartyl protease family protein] |
-0.940 |
-0.328 |
| 331 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
-0.935 |
-0.164 |
| 332 |
264404_at |
AT2G25160
|
CYP82F1, cytochrome P450, family 82, subfamily F, polypeptide 1 |
-0.923 |
0.099 |
| 333 |
248727_at |
AT5G47990
|
CYP705A5, cytochrome P450, family 705, subfamily A, polypeptide 5, THAD, THALIAN-DIOL DESATURASE, THAD1, THALIAN-DIOL DESATURASE |
-0.906 |
0.333 |
| 334 |
249567_at |
AT5G38020
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.904 |
0.587 |
| 335 |
251231_at |
AT3G62760
|
ATGSTF13 |
-0.903 |
0.046 |
| 336 |
253309_at |
AT4G33790
|
CER4, ECERIFERUM 4, FAR3, FATTY ACID REDUCTASE 3, G7 |
-0.892 |
0.078 |
| 337 |
252232_at |
AT3G49760
|
AtbZIP5, basic leucine-zipper 5, bZIP5, basic leucine-zipper 5 |
-0.891 |
-0.030 |
| 338 |
251952_at |
AT3G53650
|
[Histone superfamily protein] |
-0.890 |
-0.357 |
| 339 |
250892_at |
AT5G03760
|
ATCSLA09, ATCSLA9, CSLA09, CSLA9, CELLULOSE SYNTHASE LIKE A9, RAT4, RESISTANT TO AGROBACTERIUM TRANSFORMATION 4 |
-0.889 |
0.192 |
| 340 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.884 |
0.070 |
| 341 |
248048_at |
AT5G56080
|
ATNAS2, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 2, NAS2, nicotianamine synthase 2 |
-0.876 |
0.119 |
| 342 |
258008_at |
AT3G19430
|
[late embryogenesis abundant protein-related / LEA protein-related] |
-0.875 |
0.176 |
| 343 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
-0.875 |
-0.263 |
| 344 |
265083_at |
AT1G03820
|
unknown |
-0.867 |
-0.842 |
| 345 |
253073_at |
AT4G37410
|
CYP81F4, cytochrome P450, family 81, subfamily F, polypeptide 4 |
-0.860 |
1.031 |
| 346 |
249203_at |
AT5G42590
|
CYP71A16, cytochrome P450, family 71, subfamily A, polypeptide 16, MRO, marneral oxidase |
-0.854 |
0.231 |
| 347 |
248729_at |
AT5G48010
|
AtTHAS1, Arabidopsis thaliana thalianol synthase 1, THAS, THALIANOL SYNTHASE, THAS1, thalianol synthase 1 |
-0.847 |
0.591 |
| 348 |
247268_at |
AT5G64080
|
AtXYP1, XYP1, xylogen protein 1 |
-0.845 |
-0.302 |
| 349 |
259064_at |
AT3G07490
|
AGD11, ARF-GAP domain 11, AtCML3, CML3, calmodulin-like 3 |
-0.837 |
-0.059 |
| 350 |
259681_at |
AT1G77760
|
GNR1, NIA1, nitrate reductase 1, NR1, NITRATE REDUCTASE 1 |
-0.836 |
-0.553 |
| 351 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
-0.834 |
0.197 |
| 352 |
252858_at |
AT4G39770
|
TPPH, trehalose-6-phosphate phosphatase H |
-0.832 |
0.186 |
| 353 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
-0.832 |
0.849 |
| 354 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
-0.831 |
0.039 |
| 355 |
252501_at |
AT3G46880
|
unknown |
-0.830 |
0.084 |
| 356 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
-0.826 |
0.042 |
| 357 |
253598_at |
AT4G30800
|
[Nucleic acid-binding, OB-fold-like protein] |
-0.824 |
-0.281 |
| 358 |
248963_at |
AT5G45700
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.822 |
-0.937 |
| 359 |
247871_at |
AT5G57530
|
AtXTH12, XTH12, xyloglucan endotransglucosylase/hydrolase 12 |
-0.822 |
0.060 |
| 360 |
256368_at |
AT1G66800
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.822 |
0.055 |
| 361 |
249522_at |
AT5G38700
|
unknown |
-0.820 |
-0.242 |
| 362 |
247097_at |
AT5G66460
|
AtMAN7, MAN7, endo-beta-mannase 7 |
-0.819 |
-0.366 |
| 363 |
258618_at |
AT3G02885
|
GASA5, GAST1 protein homolog 5 |
-0.817 |
-0.463 |
| 364 |
246920_at |
AT5G25090
|
AtENODL13, ENODL13, early nodulin-like protein 13 |
-0.814 |
-0.545 |
| 365 |
264403_at |
AT2G25150
|
[HXXXD-type acyl-transferase family protein] |
-0.814 |
-0.029 |
| 366 |
261574_at |
AT1G01190
|
CYP78A8, cytochrome P450, family 78, subfamily A, polypeptide 8 |
-0.811 |
0.063 |
| 367 |
262198_at |
AT1G53830
|
ATPME2, pectin methylesterase 2, PME2, pectin methylesterase 2 |
-0.809 |
-0.017 |
| 368 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.807 |
0.170 |
| 369 |
248408_at |
AT5G51520
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.806 |
0.181 |
| 370 |
257645_at |
AT3G25790
|
[myb-like transcription factor family protein] |
-0.804 |
0.039 |
| 371 |
256878_at |
AT3G26460
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.804 |
0.100 |
| 372 |
264640_at |
AT1G65680
|
ATEXPB2, expansin B2, ATHEXP BETA 1.4, EXPB2, expansin B2 |
-0.803 |
0.148 |
| 373 |
259871_at |
AT1G76800
|
[Vacuolar iron transporter (VIT) family protein] |
-0.797 |
0.234 |
| 374 |
254371_at |
AT4G21760
|
BGLU47, beta-glucosidase 47 |
-0.795 |
0.125 |
| 375 |
245689_at |
AT5G04120
|
[Phosphoglycerate mutase family protein] |
-0.795 |
0.304 |
| 376 |
253832_at |
AT4G27654
|
unknown |
-0.792 |
0.217 |
| 377 |
255450_at |
AT4G02850
|
[phenazine biosynthesis PhzC/PhzF family protein] |
-0.791 |
0.175 |
| 378 |
267287_at |
AT2G23630
|
sks16, SKU5 similar 16 |
-0.789 |
0.047 |
| 379 |
245904_at |
AT5G11110
|
ATSPS2F, sucrose phosphate synthase 2F, KNS2, KAONASHI 2, SPS1, SUCROSE PHOSPHATE SYNTHASE 1, SPS2F, sucrose phosphate synthase 2F, SPSA2, sucrose-phosphate synthase A2 |
-0.785 |
0.054 |
| 380 |
262978_at |
AT1G75780
|
TUB1, tubulin beta-1 chain |
-0.783 |
-0.168 |
| 381 |
254534_at |
AT4G19680
|
ATIRT2, IRON REGULATED TRANSPORTER 2, IRT2, iron regulated transporter 2 |
-0.782 |
0.018 |
| 382 |
251668_at |
AT3G57010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.769 |
0.366 |
| 383 |
253696_at |
AT4G29740
|
ATCKX4, CKX4, cytokinin oxidase 4 |
-0.764 |
0.205 |
| 384 |
258859_at |
AT3G02120
|
[hydroxyproline-rich glycoprotein family protein] |
-0.762 |
-0.626 |
| 385 |
262608_at |
AT1G14120
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.761 |
1.059 |
| 386 |
248732_at |
AT5G48070
|
ATXTH20, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 20, XTH20, xyloglucan endotransglucosylase/hydrolase 20 |
-0.760 |
0.069 |
| 387 |
266828_at |
AT2G22930
|
[UDP-Glycosyltransferase superfamily protein] |
-0.745 |
0.115 |
| 388 |
267626_at |
AT2G42250
|
CYP712A1, cytochrome P450, family 712, subfamily A, polypeptide 1 |
-0.743 |
0.049 |
| 389 |
266352_at |
AT2G01610
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.741 |
-0.328 |
| 390 |
247706_at |
AT5G59480
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.740 |
0.067 |
| 391 |
245965_at |
AT5G19730
|
[Pectin lyase-like superfamily protein] |
-0.738 |
0.011 |
| 392 |
254119_at |
AT4G24780
|
[Pectin lyase-like superfamily protein] |
-0.734 |
-0.172 |
| 393 |
250651_at |
AT5G06900
|
CYP93D1, cytochrome P450, family 93, subfamily D, polypeptide 1 |
-0.733 |
0.128 |
| 394 |
247214_at |
AT5G64850
|
unknown |
-0.729 |
0.099 |
| 395 |
257735_at |
AT3G27400
|
[Pectin lyase-like superfamily protein] |
-0.728 |
-0.544 |
| 396 |
251928_at |
AT3G53980
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.724 |
0.197 |
| 397 |
256125_at |
AT1G18250
|
ATLP-1 |
-0.722 |
-0.560 |
| 398 |
251545_at |
AT3G58810
|
ATMTP3, ARABIDOPSIS METAL TOLERANCE PROTEIN 3, ATMTPA2, MTP3, METAL TOLERANCE PROTEIN 3, MTPA2, metal tolerance protein A2 |
-0.721 |
0.083 |
| 399 |
252341_at |
AT3G48940
|
[Remorin family protein] |
-0.720 |
0.153 |
| 400 |
263136_at |
AT1G78580
|
ATTPS1, trehalose-6-phosphate synthase, TPS1, trehalose-6-phosphate synthase, TPS1, TREHALOSE-6-PHOSPHATE SYNTHASE 1 |
-0.719 |
-0.025 |
| 401 |
266770_at |
AT2G03090
|
ATEXP15, ATEXPA15, expansin A15, ATHEXP ALPHA 1.3, EXP15, EXPANSIN 15, EXPA15, expansin A15 |
-0.719 |
0.087 |
| 402 |
266996_at |
AT2G34490
|
CYP710A2, cytochrome P450, family 710, subfamily A, polypeptide 2 |
-0.716 |
0.438 |
| 403 |
246403_at |
AT1G57590
|
[Pectinacetylesterase family protein] |
-0.713 |
0.262 |
| 404 |
249063_at |
AT5G44110
|
ABCI21, ATP-binding cassette A21, ATNAP2, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN, ATPOP1, POP1 |
-0.710 |
-0.015 |
| 405 |
258396_at |
AT3G15460
|
[Ribosomal RNA processing Brix domain protein] |
-0.708 |
-0.324 |
| 406 |
264672_at |
AT1G09750
|
[Eukaryotic aspartyl protease family protein] |
-0.705 |
-0.283 |
| 407 |
255645_at |
AT4G00880
|
SAUR31, SMALL AUXIN UPREGULATED RNA 31 |
-0.705 |
0.002 |
| 408 |
254632_at |
AT4G18630
|
unknown |
-0.704 |
-0.739 |
| 409 |
255773_at |
AT1G18590
|
ATSOT17, SULFOTRANSFERASE 17, ATST5C, ARABIDOPSIS SULFOTRANSFERASE 5C, SOT17, sulfotransferase 17 |
-0.704 |
0.446 |
| 410 |
263594_at |
AT2G01880
|
ATPAP7, PURPLE ACID PHOSPHATASE 7, PAP7, purple acid phosphatase 7 |
-0.703 |
0.468 |
| 411 |
248681_at |
AT5G48900
|
[Pectin lyase-like superfamily protein] |
-0.703 |
-0.078 |
| 412 |
255818_at |
AT2G33570
|
GALS1, galactan synthase 1 |
-0.702 |
-0.209 |
| 413 |
246395_at |
AT1G58170
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.700 |
0.218 |
| 414 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
-0.699 |
-0.025 |
| 415 |
253017_at |
AT4G37970
|
ATCAD6, CAD6, cinnamyl alcohol dehydrogenase 6 |
-0.696 |
0.460 |
| 416 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
-0.692 |
0.157 |
| 417 |
252202_at |
AT3G50300
|
[HXXXD-type acyl-transferase family protein] |
-0.689 |
0.164 |
| 418 |
266790_at |
AT2G28950
|
ATEXP6, ARABIDOPSIS THALIANA TEXPANSIN 6, ATEXPA6, expansin A6, ATHEXP ALPHA 1.8, EXPA6, expansin A6 |
-0.688 |
-0.657 |
| 419 |
248684_at |
AT5G48485
|
DIR1, DEFECTIVE IN INDUCED RESISTANCE 1 |
-0.687 |
-0.112 |
| 420 |
264909_at |
AT2G17300
|
unknown |
-0.686 |
-0.352 |
| 421 |
248725_at |
AT5G47980
|
[HXXXD-type acyl-transferase family protein] |
-0.683 |
0.240 |
| 422 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.683 |
-0.021 |
| 423 |
250434_at |
AT5G10390
|
H3.1, histone 3.1 |
-0.682 |
-0.159 |
| 424 |
266418_at |
AT2G38750
|
ANNAT4, annexin 4, AtANN4 |
-0.681 |
0.976 |
| 425 |
258912_at |
AT3G06460
|
[GNS1/SUR4 membrane protein family] |
-0.681 |
0.074 |
| 426 |
245967_at |
AT5G19800
|
[hydroxyproline-rich glycoprotein family protein] |
-0.681 |
0.180 |
| 427 |
247476_at |
AT5G62330
|
unknown |
-0.678 |
0.062 |
| 428 |
258675_at |
AT3G08770
|
LTP6, lipid transfer protein 6 |
-0.677 |
0.037 |
| 429 |
254837_at |
AT4G12360
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.677 |
0.098 |
| 430 |
259655_at |
AT1G55210
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.675 |
-0.021 |
| 431 |
262231_at |
AT1G68740
|
PHO1;H1 |
-0.675 |
0.199 |
| 432 |
255284_at |
AT4G04610
|
APR, APR1, APS reductase 1, ATAPR1, PRH19, PAPS REDUCTASE HOMOLOG 19 |
-0.673 |
-0.205 |
| 433 |
256600_at |
AT3G14850
|
TBL41, TRICHOME BIREFRINGENCE-LIKE 41 |
-0.672 |
-0.015 |
| 434 |
253515_at |
AT4G31320
|
SAUR37, SMALL AUXIN UPREGULATED RNA 37 |
-0.671 |
0.090 |
| 435 |
266271_at |
AT2G29440
|
ATGSTU6, glutathione S-transferase tau 6, GST24, GLUTATHIONE S-TRANSFERASE 24, GSTU6, glutathione S-transferase tau 6 |
-0.670 |
0.256 |
| 436 |
266765_at |
AT2G46860
|
AtPPa3, pyrophosphorylase 3, PPa3, pyrophosphorylase 3 |
-0.670 |
0.122 |
| 437 |
263841_at |
AT2G36870
|
AtXTH32, XTH32, xyloglucan endotransglucosylase/hydrolase 32 |
-0.670 |
0.185 |
| 438 |
253949_at |
AT4G26780
|
AR192, MGE2, mitochondrial GrpE 2 |
-0.667 |
-0.170 |
| 439 |
263553_at |
AT2G16430
|
ATPAP10, PAP10, purple acid phosphatase 10 |
-0.667 |
0.013 |
| 440 |
249756_at |
AT5G24313
|
unknown |
-0.665 |
0.036 |
| 441 |
262109_at |
AT1G02730
|
ATCSLD5, cellulose synthase-like D5, CSLD5, CELLULOSE SYNTHASE LIKE D5, CSLD5, cellulose synthase-like D5, SOS6, SALT OVERLY SENSITIVE 6 |
-0.663 |
-0.586 |
| 442 |
249773_at |
AT5G24140
|
SQP2, squalene monooxygenase 2 |
-0.662 |
0.443 |
| 443 |
253088_at |
AT4G36220
|
CYP84A1, CYTOCHROME P450 84A1, FAH1, ferulic acid 5-hydroxylase 1 |
-0.662 |
0.394 |
| 444 |
255460_at |
AT4G02800
|
unknown |
-0.661 |
-0.774 |
| 445 |
257204_at |
AT3G23805
|
RALFL24, ralf-like 24 |
-0.661 |
-0.071 |
| 446 |
254326_at |
AT4G22610
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.658 |
0.828 |
| 447 |
249893_at |
AT5G22555
|
unknown |
-0.658 |
0.036 |
| 448 |
259850_at |
AT1G72240
|
unknown |
-0.657 |
0.030 |
| 449 |
253010_at |
AT4G37750
|
ANT, AINTEGUMENTA, CKC, CKC1, COMPLEMENTING A PROTEIN KINASE C MUTANT 1, DRG, DRAGON |
-0.657 |
-0.293 |
| 450 |
249202_at |
AT5G42580
|
CYP705A12, cytochrome P450, family 705, subfamily A, polypeptide 12 |
-0.657 |
-0.014 |
| 451 |
247914_at |
AT5G57540
|
AtXTH13, XTH13, xyloglucan endotransglucosylase/hydrolase 13 |
-0.656 |
0.163 |
| 452 |
257203_at |
AT3G23730
|
XTH16, xyloglucan endotransglucosylase/hydrolase 16 |
-0.655 |
0.032 |
| 453 |
259763_at |
AT1G77630
|
LYM3, lysin-motif (LysM) domain protein 3, LYP3, LysM-containing receptor protein 3 |
-0.655 |
-0.164 |
| 454 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-0.654 |
0.110 |
| 455 |
257977_at |
AT3G20850
|
[proline-rich family protein] |
-0.653 |
0.077 |
| 456 |
257668_at |
AT3G20460
|
[Major facilitator superfamily protein] |
-0.652 |
0.093 |
| 457 |
251287_at |
AT3G61820
|
[Eukaryotic aspartyl protease family protein] |
-0.651 |
-0.235 |
| 458 |
260059_at |
AT1G78090
|
ATTPPB, Arabidopsis thaliana trehalose-6-phosphate phosphatase B, TPPB, trehalose-6-phosphate phosphatase B |
-0.650 |
0.101 |
| 459 |
260119_at |
AT1G33930
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.649 |
0.026 |
| 460 |
259801_at |
AT1G72230
|
[Cupredoxin superfamily protein] |
-0.648 |
0.258 |
| 461 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
-0.648 |
-0.073 |
| 462 |
250173_at |
AT5G14340
|
AtMYB40, myb domain protein 40, MYB40, myb domain protein 40 |
-0.647 |
0.091 |
| 463 |
265463_at |
AT2G37090
|
IRX9, IRREGULAR XYLEM 9 |
-0.647 |
0.119 |
| 464 |
263376_at |
AT2G20520
|
FLA6, FASCICLIN-like arabinogalactan 6 |
-0.647 |
-0.021 |
| 465 |
254332_at |
AT4G22730
|
[Leucine-rich repeat protein kinase family protein] |
-0.647 |
-0.067 |
| 466 |
256073_at |
AT1G18100
|
E12A11, MFT, MOTHER OF FT AND TFL1 |
-0.643 |
0.566 |
| 467 |
249340_at |
AT5G41140
|
[Myosin heavy chain-related protein] |
-0.643 |
-0.199 |
| 468 |
267137_at |
AT2G23410
|
ACPT, cis-prenyltransferase, AtcPT3, CPT, cis-prenyltransferase, cPT3, cis-prenyltransferase 3 |
-0.643 |
-0.015 |
| 469 |
249971_at |
AT5G19110
|
[Eukaryotic aspartyl protease family protein] |
-0.640 |
0.834 |
| 470 |
256319_at |
AT1G35910
|
TPPD, trehalose-6-phosphate phosphatase D |
-0.640 |
0.411 |
| 471 |
256352_at |
AT1G54970
|
ATPRP1, proline-rich protein 1, PRP1, proline-rich protein 1, RHS7, ROOT HAIR SPECIFIC 7 |
-0.639 |
0.138 |
| 472 |
259466_at |
AT1G19050
|
ARR7, response regulator 7 |
-0.635 |
-0.133 |
| 473 |
264565_at |
AT1G05280
|
unknown |
-0.635 |
0.204 |
| 474 |
260547_at |
AT2G43550
|
[Scorpion toxin-like knottin superfamily protein] |
-0.628 |
0.531 |
| 475 |
247030_at |
AT5G67210
|
IRX15-L, IRX15-LIKE |
-0.628 |
0.022 |
| 476 |
254107_at |
AT4G25220
|
AtG3Pp2, glycerol-3-phosphate permease 2, G3Pp2, glycerol-3-phosphate permease 2, RHS15, root hair specific 15 |
-0.628 |
0.123 |
| 477 |
250933_at |
AT5G03170
|
ATFLA11, ARABIDOPSIS FASCICLIN-LIKE ARABINOGALACTAN-PROTEIN 11, FLA11, FASCICLIN-like arabinogalactan-protein 11 |
-0.627 |
0.045 |
| 478 |
250469_at |
AT5G10130
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.626 |
0.150 |
| 479 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
-0.625 |
0.374 |
| 480 |
264511_at |
AT1G09350
|
AtGolS3, galactinol synthase 3, GolS3, galactinol synthase 3 |
-0.625 |
0.102 |
| 481 |
254634_at |
AT4G18650
|
[transcription factor-related] |
-0.625 |
0.163 |
| 482 |
261133_at |
AT1G19715
|
[Mannose-binding lectin superfamily protein] |
-0.624 |
-0.002 |
| 483 |
249832_at |
AT5G23400
|
[Leucine-rich repeat (LRR) family protein] |
-0.623 |
-0.500 |
| 484 |
250337_at |
AT5G11790
|
NDL2, N-MYC downregulated-like 2 |
-0.623 |
-0.131 |
| 485 |
263942_at |
AT2G35860
|
FLA16, FASCICLIN-like arabinogalactan protein 16 precursor |
-0.622 |
-0.360 |
| 486 |
253255_at |
AT4G34760
|
SAUR50, SMALL AUXIN UPREGULATED RNA 50 |
-0.621 |
0.018 |
| 487 |
255876_at |
AT2G40480
|
unknown |
-0.620 |
-0.171 |
| 488 |
253687_at |
AT4G29520
|
[LOCATED IN: endoplasmic reticulum, plasma membrane] |
-0.620 |
-0.109 |
| 489 |
262795_at |
AT1G13130
|
[Cellulase (glycosyl hydrolase family 5) protein] |
-0.618 |
0.064 |
| 490 |
264029_at |
AT2G03720
|
MRH6, morphogenesis of root hair 6 |
-0.618 |
0.067 |
| 491 |
252763_at |
AT3G42725
|
[Putative membrane lipoprotein] |
-0.618 |
-0.827 |
| 492 |
259346_at |
AT3G03910
|
glutamate dehydrogenase 3 |
-0.617 |
0.137 |
| 493 |
264262_at |
AT1G09200
|
H3.1, histone 3.1 |
-0.617 |
-0.198 |
| 494 |
262659_at |
AT1G14240
|
[GDA1/CD39 nucleoside phosphatase family protein] |
-0.616 |
0.964 |
| 495 |
261840_at |
AT1G16070
|
AtTLP8, tubby like protein 8, TLP8, tubby like protein 8 |
-0.610 |
-0.672 |
| 496 |
263677_at |
AT1G04520
|
PDLP2, plasmodesmata-located protein 2 |
-0.610 |
-0.304 |
| 497 |
263452_at |
AT2G22190
|
TPPE, trehalose-6-phosphate phosphatase E |
-0.610 |
0.130 |
| 498 |
246617_at |
AT5G36270
|
[similar to DHAR2, glutathione dehydrogenase (ascorbate) [Arabidopsis thaliana] (TAIR:AT1G75270.1)] |
-0.609 |
0.024 |
| 499 |
261921_at |
AT1G65900
|
unknown |
-0.609 |
0.092 |
| 500 |
264573_at |
AT1G05310
|
[Pectin lyase-like superfamily protein] |
-0.606 |
0.002 |
| 501 |
249046_at |
AT5G44400
|
[FAD-binding Berberine family protein] |
-0.605 |
-0.653 |
| 502 |
257151_at |
AT3G27200
|
[Cupredoxin superfamily protein] |
-0.604 |
0.090 |
| 503 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
-0.602 |
-0.087 |
| 504 |
249718_at |
AT5G35740
|
[Carbohydrate-binding X8 domain superfamily protein] |
-0.601 |
0.121 |
| 505 |
264391_at |
AT1G11920
|
[Pectin lyase-like superfamily protein] |
-0.601 |
0.090 |
| 506 |
252594_at |
AT3G45680
|
[Major facilitator superfamily protein] |
-0.599 |
0.184 |
| 507 |
259195_at |
AT3G01730
|
unknown |
-0.596 |
0.173 |
| 508 |
246530_at |
AT5G15725
|
GLV9, GOLVEN 9 |
-0.596 |
-0.019 |
| 509 |
256128_at |
AT1G18140
|
ATLAC1, LAC1, laccase 1 |
-0.595 |
0.178 |
| 510 |
259878_at |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
-0.591 |
0.999 |
| 511 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
-0.588 |
-0.003 |
| 512 |
258938_at |
AT3G10080
|
[RmlC-like cupins superfamily protein] |
-0.588 |
0.249 |
| 513 |
261203_at |
AT1G12845
|
unknown |
-0.588 |
0.140 |
| 514 |
256093_at |
AT1G20823
|
[RING/U-box superfamily protein] |
-0.587 |
-0.136 |
| 515 |
261448_at |
AT1G21140
|
[Vacuolar iron transporter (VIT) family protein] |
-0.586 |
0.066 |
| 516 |
246827_at |
AT5G26330
|
[Cupredoxin superfamily protein] |
-0.586 |
-0.009 |
| 517 |
247962_at |
AT5G56580
|
ANQ1, ARABIDOPSIS NQK1, ATMKK6, ARABIDOPSIS THALIANA MAP KINASE KINASE 6, MKK6, MAP kinase kinase 6 |
-0.585 |
-0.703 |
| 518 |
258178_at |
AT3G21680
|
unknown |
-0.584 |
0.067 |
| 519 |
245546_at |
AT4G15290
|
ATCSLB05, ATCSLB5, CELLULOSE SYNTHASE LIKE 5, CSLB05 |
-0.583 |
0.105 |
| 520 |
249920_at |
AT5G19260
|
FAF3, FANTASTIC FOUR 3 |
-0.583 |
0.197 |
| 521 |
252442_at |
AT3G46940
|
DUT1, DUTP-PYROPHOSPHATASE-LIKE 1 |
-0.582 |
-0.235 |
| 522 |
264347_at |
AT1G12040
|
LRX1, leucine-rich repeat/extensin 1 |
-0.581 |
-0.043 |
| 523 |
257714_at |
AT3G27360
|
H3.1, histone 3.1 |
-0.581 |
-0.222 |
| 524 |
264315_at |
AT1G70370
|
PG2, polygalacturonase 2 |
-0.580 |
-0.234 |
| 525 |
261541_at |
AT1G63600
|
[Receptor-like protein kinase-related family protein] |
-0.579 |
0.096 |
| 526 |
249881_at |
AT5G23190
|
CYP86B1, cytochrome P450, family 86, subfamily B, polypeptide 1 |
-0.579 |
0.106 |
| 527 |
266684_at |
AT2G19720
|
rps15ab, ribosomal protein S15A B |
-0.579 |
-0.081 |
| 528 |
260150_at |
AT1G52820
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.578 |
0.098 |
| 529 |
253090_at |
AT4G36360
|
BGAL3, beta-galactosidase 3 |
-0.577 |
-0.430 |
| 530 |
252302_at |
AT3G49190
|
[O-acyltransferase (WSD1-like) family protein] |
-0.577 |
0.062 |
| 531 |
247727_at |
AT5G59490
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.576 |
0.368 |
| 532 |
266253_at |
AT2G27840
|
HD2D, histone deacetylase 2D, HDA13, HISTONE DEACETYLASE 13, HDT04, HDT4 |
-0.574 |
-0.282 |
| 533 |
262148_at |
AT1G52560
|
[HSP20-like chaperones superfamily protein] |
-0.574 |
0.280 |
| 534 |
266413_at |
AT2G38740
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.574 |
0.209 |
| 535 |
249868_at |
AT5G23030
|
TET12, tetraspanin12 |
-0.572 |
0.130 |
| 536 |
253720_at |
AT4G29270
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.572 |
0.544 |
| 537 |
267411_at |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
-0.571 |
0.361 |
| 538 |
248891_at |
AT5G46280
|
MCM3, MINICHROMOSOME MAINTENANCE 3 |
-0.570 |
-0.271 |
| 539 |
259516_at |
AT1G20450
|
ERD10, EARLY RESPONSIVE TO DEHYDRATION 10, LTI29, LOW TEMPERATURE INDUCED 29, LTI45, LOW TEMPERATURE INDUCED 45 |
-0.570 |
0.002 |
| 540 |
245739_at |
AT1G44110
|
CYCA1;1, Cyclin A1;1 |
-0.569 |
-0.578 |
| 541 |
261080_at |
AT1G07370
|
ATPCNA1, PROLIFERATING CELLULAR NUCLEAR ANTIGEN 1, PCNA1, proliferating cellular nuclear antigen 1 |
-0.568 |
-0.259 |
| 542 |
257428_at |
AT1G78990
|
[HXXXD-type acyl-transferase family protein] |
-0.567 |
-0.028 |
| 543 |
264287_at |
AT1G61930
|
unknown |
-0.567 |
-0.117 |
| 544 |
252089_at |
AT3G52110
|
unknown |
-0.565 |
-0.571 |
| 545 |
248150_at |
AT5G54670
|
ATK3, kinesin 3, KATC, KINESIN-LIKE PROTEIN IN ARABIDOPSIS THALIANA C |
-0.565 |
-0.234 |
| 546 |
257815_at |
AT3G25130
|
unknown |
-0.561 |
-0.628 |
| 547 |
261099_at |
AT1G62980
|
ATEXP18, ATEXPA18, expansin A18, ATHEXP ALPHA 1.25, EXP18, EXPANSIN 18, EXPA18, expansin A18 |
-0.560 |
0.058 |
| 548 |
249797_at |
AT5G23750
|
[Remorin family protein] |
-0.558 |
0.343 |
| 549 |
245343_at |
AT4G15830
|
[ARM repeat superfamily protein] |
-0.556 |
-0.660 |
| 550 |
265117_at |
AT1G62500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.554 |
0.140 |
| 551 |
264010_at |
AT2G21100
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.553 |
0.128 |
| 552 |
261417_at |
AT1G07700
|
[Thioredoxin superfamily protein] |
-0.552 |
0.102 |
| 553 |
258529_at |
AT3G06740
|
GATA15, GATA transcription factor 15 |
-0.552 |
-0.282 |
| 554 |
255011_at |
AT4G10040
|
CYTC-2, cytochrome c-2 |
-0.551 |
0.032 |
| 555 |
249392_at |
AT5G40150
|
[Peroxidase superfamily protein] |
-0.550 |
-0.497 |
| 556 |
262972_at |
AT1G75620
|
[glyoxal oxidase-related protein] |
-0.549 |
0.070 |
| 557 |
251067_at |
AT5G01910
|
unknown |
-0.549 |
-0.642 |
| 558 |
266170_at |
AT2G39050
|
ArathEULS3, EULS3, Euonymus lectin S3 |
-0.549 |
0.390 |
| 559 |
246557_at |
AT5G15510
|
[TPX2 (targeting protein for Xklp2) protein family] |
-0.549 |
-0.302 |
| 560 |
258613_at |
AT3G02870
|
VTC4 |
-0.548 |
0.007 |
| 561 |
249583_at |
AT5G37770
|
CML24, CALMODULIN-LIKE 24, TCH2, TOUCH 2 |
-0.548 |
-0.249 |
| 562 |
265204_at |
AT2G36650
|
unknown |
-0.547 |
0.024 |
| 563 |
254313_at |
AT4G22460
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.546 |
0.144 |
| 564 |
256563_at |
AT3G29780
|
RALFL27, ralf-like 27 |
-0.545 |
0.055 |
| 565 |
252711_at |
AT3G43720
|
LTPG2, glycosylphosphatidylinositol-anchored lipid protein transfer 2 |
-0.545 |
0.014 |
| 566 |
251584_at |
AT3G58620
|
TTL4, tetratricopetide-repeat thioredoxin-like 4 |
-0.542 |
0.046 |
| 567 |
261881_at |
AT1G80760
|
NIP6, NIP6;1, NOD26-like intrinsic protein 6;1, NLM7 |
-0.542 |
0.002 |
| 568 |
255345_at |
AT4G04460
|
[Saposin-like aspartyl protease family protein] |
-0.541 |
0.445 |
| 569 |
253165_at |
AT4G35320
|
unknown |
-0.540 |
-0.286 |
| 570 |
261399_at |
AT1G79620
|
[Leucine-rich repeat protein kinase family protein] |
-0.540 |
0.187 |
| 571 |
258470_at |
AT3G06035
|
[Glycoprotein membrane precursor GPI-anchored] |
-0.539 |
0.082 |
| 572 |
259632_at |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
-0.537 |
0.130 |
| 573 |
247406_at |
AT5G62920
|
ARR6, response regulator 6 |
-0.537 |
0.064 |
| 574 |
266089_at |
AT2G38010
|
[Neutral/alkaline non-lysosomal ceramidase] |
-0.536 |
-0.032 |
| 575 |
250230_at |
AT5G13900
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.536 |
-0.013 |
| 576 |
258551_at |
AT3G06890
|
unknown |
-0.535 |
-0.183 |
| 577 |
245698_at |
AT5G04160
|
[Nucleotide-sugar transporter family protein] |
-0.534 |
-0.148 |
| 578 |
257824_at |
AT3G25290
|
[Auxin-responsive family protein] |
-0.533 |
-0.236 |
| 579 |
255104_at |
AT4G08685
|
SAH7 |
-0.532 |
-0.004 |
| 580 |
247037_at |
AT5G67070
|
RALFL34, ralf-like 34 |
-0.532 |
-0.197 |
| 581 |
250958_at |
AT5G03260
|
LAC11, laccase 11 |
-0.532 |
0.044 |
| 582 |
254888_at |
AT4G11780
|
TRM10, TON1 Recruiting Motif 10 |
-0.532 |
0.089 |
| 583 |
253480_at |
AT4G31840
|
AtENODL15, ENODL15, early nodulin-like protein 15 |
-0.531 |
-0.548 |
| 584 |
245966_at |
AT5G19790
|
RAP2.11, related to AP2 11 |
-0.529 |
0.121 |
| 585 |
257481_at |
AT1G08430
|
ALMT1, aluminum-activated malate transporter 1, ATALMT1, ARABIDOPSIS THALIANA ALUMINUM-ACTIVATED MALATE TRANSPORTER 1 |
-0.528 |
0.130 |
| 586 |
258765_at |
AT3G10710
|
RHS12, root hair specific 12 |
-0.528 |
-0.008 |
| 587 |
264184_at |
AT1G54790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.525 |
0.103 |
| 588 |
255517_at |
AT4G02290
|
AtGH9B13, glycosyl hydrolase 9B13, GH9B13, glycosyl hydrolase 9B13 |
-0.525 |
-0.484 |
| 589 |
264968_at |
AT1G67360
|
[Rubber elongation factor protein (REF)] |
-0.525 |
0.076 |
| 590 |
249576_at |
AT5G37690
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.525 |
0.076 |
| 591 |
253286_at |
AT4G34260
|
AXY8, ALTERED XYLOGLUCAN 8, FUC95A |
-0.523 |
-0.149 |
| 592 |
258222_at |
AT3G15680
|
[Ran BP2/NZF zinc finger-like superfamily protein] |
-0.523 |
0.180 |
| 593 |
256926_at |
AT3G22540
|
unknown |
-0.522 |
-0.776 |
| 594 |
259846_at |
AT1G72140
|
[Major facilitator superfamily protein] |
-0.521 |
0.414 |
| 595 |
257512_at |
AT1G35250
|
ALT2, acyl-lipid thioesterase 2 |
-0.520 |
0.066 |
| 596 |
265155_at |
AT1G30990
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.520 |
0.008 |
| 597 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
-0.520 |
-0.429 |
| 598 |
249214_at |
AT5G42720
|
[Glycosyl hydrolase family 17 protein] |
-0.519 |
-0.009 |
| 599 |
260135_at |
AT1G66400
|
CML23, calmodulin like 23 |
-0.519 |
0.057 |
| 600 |
251062_at |
AT5G01840
|
ATOFP1, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 1, OFP1, ovate family protein 1 |
-0.519 |
0.368 |