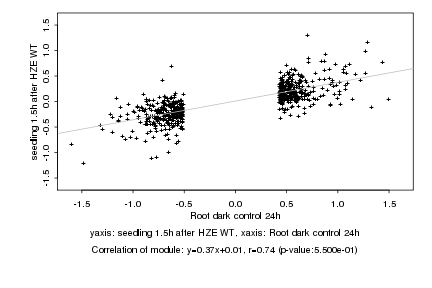
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Root dark control 24h |
Log2 signal ratio
seedling 1.5h after HZE WT |
| 1 |
262325_at |
AT1G64160
|
AtDIR5, DIR5, dirigent protein 5 |
1.494 |
0.048 |
| 2 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
1.436 |
0.778 |
| 3 |
249800_at |
AT5G23660
|
AtSWEET12, MTN3, homolog of Medicago truncatula MTN3, SWEET12 |
1.322 |
-0.106 |
| 4 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
1.284 |
1.173 |
| 5 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
1.269 |
0.565 |
| 6 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
1.267 |
0.995 |
| 7 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
1.212 |
0.424 |
| 8 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
1.164 |
0.542 |
| 9 |
267262_at |
AT2G22990
|
SCPL8, SERINE CARBOXYPEPTIDASE-LIKE 8, SNG1, sinapoylglucose 1 |
1.142 |
0.054 |
| 10 |
257832_at |
AT3G26740
|
CCL, CCR-like |
1.106 |
0.728 |
| 11 |
264774_at |
AT1G22890
|
unknown |
1.087 |
0.356 |
| 12 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
1.076 |
0.558 |
| 13 |
253322_at |
AT4G33980
|
unknown |
1.073 |
0.246 |
| 14 |
245528_at |
AT4G15530
|
PPDK, pyruvate orthophosphate dikinase |
1.070 |
0.687 |
| 15 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
1.067 |
0.318 |
| 16 |
265481_at |
AT2G15960
|
unknown |
1.055 |
0.631 |
| 17 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
1.053 |
0.573 |
| 18 |
258983_at |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
1.024 |
-0.052 |
| 19 |
250425_at |
AT5G10530
|
LecRK-IX.1, L-type lectin receptor kinase IX.1 |
1.012 |
0.151 |
| 20 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
1.011 |
0.374 |
| 21 |
263126_at |
AT1G78460
|
[SOUL heme-binding family protein] |
1.010 |
0.212 |
| 22 |
258923_at |
AT3G10450
|
SCPL7, serine carboxypeptidase-like 7 |
0.995 |
0.060 |
| 23 |
259244_at |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
0.978 |
0.316 |
| 24 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
0.972 |
0.732 |
| 25 |
263198_at |
AT1G53990
|
GLIP3, GDSL-motif lipase 3 |
0.963 |
0.149 |
| 26 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
0.949 |
0.313 |
| 27 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.946 |
0.431 |
| 28 |
261558_at |
AT1G01770
|
unknown |
0.946 |
0.390 |
| 29 |
265998_at |
AT2G24270
|
ALDH11A3, aldehyde dehydrogenase 11A3 |
0.926 |
-0.052 |
| 30 |
252327_at |
AT3G48740
|
AtSWEET11, SWEET11 |
0.926 |
-0.063 |
| 31 |
259316_at |
AT3G01175
|
unknown |
0.920 |
0.348 |
| 32 |
250868_at |
AT5G03860
|
MLS, malate synthase |
0.914 |
0.453 |
| 33 |
258395_at |
AT3G15500
|
ANAC055, NAC domain containing protein 55, ATNAC3, NAC domain containing protein 3, NAC055, NAC domain containing protein 55, NAC3, NAC domain containing protein 3 |
0.909 |
0.632 |
| 34 |
250940_at |
AT5G03310
|
SAUR44, SMALL AUXIN UPREGULATED RNA 44 |
0.893 |
0.228 |
| 35 |
262580_at |
AT1G15330
|
[Cystathionine beta-synthase (CBS) protein] |
0.873 |
0.939 |
| 36 |
250649_at |
AT5G06690
|
WCRKC1, WCRKC thioredoxin 1 |
0.865 |
0.787 |
| 37 |
258338_at |
AT3G16150
|
ASPGB1, asparaginase B1 |
0.863 |
0.445 |
| 38 |
264482_at |
AT1G77210
|
AtSTP14, sugar transport protein 14, STP14, sugar transport protein 14 |
0.862 |
0.627 |
| 39 |
260522_x_at |
AT2G41730
|
unknown |
0.858 |
0.121 |
| 40 |
258383_at |
AT3G15440
|
unknown |
0.847 |
0.260 |
| 41 |
259982_at |
AT1G76410
|
ATL8 |
0.839 |
0.791 |
| 42 |
248931_at |
AT5G46040
|
[Major facilitator superfamily protein] |
0.827 |
0.109 |
| 43 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
0.814 |
0.450 |
| 44 |
260795_at |
AT1G06225
|
CLE3, CLAVATA3/ESR-RELATED 3 |
0.797 |
0.052 |
| 45 |
265070_at |
AT1G55510
|
BCDH BETA1, branched-chain alpha-keto acid decarboxylase E1 beta subunit |
0.775 |
0.561 |
| 46 |
245123_at |
AT2G47450
|
CAO, CHAOS, CPSRP43, CHLOROPLAST SIGNAL RECOGNITION PARTICLE 43 |
0.774 |
0.044 |
| 47 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.762 |
-0.045 |
| 48 |
261572_at |
AT1G01170
|
unknown |
0.762 |
0.199 |
| 49 |
255893_at |
AT1G17960
|
[Threonyl-tRNA synthetase] |
0.757 |
0.283 |
| 50 |
266355_at |
AT2G01400
|
unknown |
0.757 |
0.202 |
| 51 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
0.757 |
0.072 |
| 52 |
259694_at |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
0.748 |
0.411 |
| 53 |
248744_at |
AT5G48250
|
BBX8, B-box domain protein 8 |
0.731 |
0.195 |
| 54 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
0.729 |
-0.094 |
| 55 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
0.715 |
0.036 |
| 56 |
260662_at |
AT1G19540
|
[NmrA-like negative transcriptional regulator family protein] |
0.711 |
0.856 |
| 57 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.707 |
0.132 |
| 58 |
254001_at |
AT4G26260
|
MIOX4, myo-inositol oxygenase 4 |
0.706 |
0.772 |
| 59 |
256698_at |
AT3G20680
|
unknown |
0.706 |
0.142 |
| 60 |
261821_at |
AT1G11530
|
ATCXXS1, C-terminal cysteine residue is changed to a serine 1, CXXS1, C-terminal cysteine residue is changed to a serine 1 |
0.705 |
0.301 |
| 61 |
260529_at |
AT2G47400
|
CP12, CP12 DOMAIN-CONTAINING PROTEIN 1, CP12-1, CP12 domain-containing protein 1 |
0.696 |
0.049 |
| 62 |
262322_at |
AT1G27590
|
unknown |
0.696 |
0.145 |
| 63 |
252539_at |
AT3G45730
|
unknown |
0.696 |
1.311 |
| 64 |
246831_at |
AT5G26340
|
ATSTP13, SUGAR TRANSPORT PROTEIN 13, MSS1, STP13, SUGAR TRANSPORT PROTEIN 13 |
0.693 |
0.123 |
| 65 |
248050_at |
AT5G56100
|
[glycine-rich protein / oleosin] |
0.687 |
0.382 |
| 66 |
248735_at |
AT5G48100
|
ATLAC15, AtTT10, LAC15, LACCASE-LIKE 15, TT10, TRANSPARENT TESTA 10 |
0.681 |
0.151 |
| 67 |
262635_at |
AT1G06570
|
HPD, 4-hydroxyphenylpyruvate dioxygenase, HPPD, 4-hydroxyphenylpyruvate dioxygenase, PDS1, phytoene desaturation 1 |
0.681 |
0.421 |
| 68 |
246432_at |
AT5G17490
|
AtRGL3, RGL3, RGA-like protein 3 |
0.680 |
0.223 |
| 69 |
256340_at |
AT1G72070
|
[Chaperone DnaJ-domain superfamily protein] |
0.674 |
0.175 |
| 70 |
250772_at |
AT5G05420
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
0.672 |
0.050 |
| 71 |
261618_at |
AT1G33110
|
[MATE efflux family protein] |
0.670 |
-0.222 |
| 72 |
263556_at |
AT2G16365
|
[F-box family protein] |
0.669 |
-0.121 |
| 73 |
247326_at |
AT5G64110
|
[Peroxidase superfamily protein] |
0.665 |
0.105 |
| 74 |
257615_at |
AT3G26510
|
[Octicosapeptide/Phox/Bem1p family protein] |
0.654 |
0.165 |
| 75 |
258527_at |
AT3G06850
|
BCE2, DIN3, DARK INDUCIBLE 3, LTA1 |
0.654 |
0.507 |
| 76 |
260741_at |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
0.653 |
0.473 |
| 77 |
248975_at |
AT5G45040
|
CYTC6A, cytochrome c6A |
0.652 |
0.196 |
| 78 |
260014_at |
AT1G68010
|
ATHPR1, HPR, hydroxypyruvate reductase |
0.651 |
0.043 |
| 79 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
0.650 |
0.303 |
| 80 |
259441_at |
AT1G02300
|
[Cysteine proteinases superfamily protein] |
0.645 |
0.506 |
| 81 |
266925_at |
AT2G45740
|
PEX11D, peroxin 11D |
0.643 |
0.169 |
| 82 |
248347_at |
AT5G52250
|
EFO1, EARLY FLOWERING BY OVEREXPRESSION 1, RUP1, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 |
0.642 |
0.246 |
| 83 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
0.640 |
0.166 |
| 84 |
246951_at |
AT5G04880
|
[expressed protein] |
0.640 |
0.527 |
| 85 |
260411_at |
AT1G69890
|
unknown |
0.640 |
0.411 |
| 86 |
265511_at |
AT2G05540
|
[Glycine-rich protein family] |
0.637 |
0.109 |
| 87 |
261613_at |
AT1G49720
|
ABF1, abscisic acid responsive element-binding factor 1, AtABF1 |
0.630 |
0.508 |
| 88 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
0.630 |
0.206 |
| 89 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.629 |
0.421 |
| 90 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
0.625 |
0.355 |
| 91 |
265149_at |
AT1G51400
|
[Photosystem II 5 kD protein] |
0.624 |
0.064 |
| 92 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
0.622 |
0.300 |
| 93 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
0.619 |
0.374 |
| 94 |
249905_at |
AT5G22720
|
[F-box/RNI-like superfamily protein] |
0.619 |
0.271 |
| 95 |
262546_at |
AT1G31260
|
ZIP10, zinc transporter 10 precursor |
0.619 |
0.182 |
| 96 |
261211_at |
AT1G12780
|
ATUGE1, A. THALIANA UDP-GLC 4-EPIMERASE 1, UGE1, UDP-D-glucose/UDP-D-galactose 4-epimerase 1 |
0.617 |
0.427 |
| 97 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
0.615 |
0.366 |
| 98 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
0.613 |
0.549 |
| 99 |
264892_at |
AT1G23160
|
[Auxin-responsive GH3 family protein] |
0.611 |
-0.277 |
| 100 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.609 |
-0.084 |
| 101 |
251497_at |
AT3G59060
|
PIF5, PHYTOCHROME-INTERACTING FACTOR 5, PIL6, phytochrome interacting factor 3-like 6 |
0.609 |
0.098 |
| 102 |
262629_at |
AT1G06460
|
ACD31.2, ALPHA-CRYSTALLIN DOMAIN 31.2, ACD32.1, alpha-crystallin domain 32.1 |
0.605 |
0.065 |
| 103 |
265992_at |
AT2G24130
|
[Leucine-rich receptor-like protein kinase family protein] |
0.605 |
0.025 |
| 104 |
251082_at |
AT5G01530
|
LHCB4.1, light harvesting complex photosystem II |
0.603 |
0.052 |
| 105 |
263350_at |
AT2G13360
|
AGT, alanine:glyoxylate aminotransferase, AGT1, ALANINE:GLYOXYLATE AMINOTRANSFERASE 1, SGAT, L-serine:glyoxylate aminotransferase |
0.601 |
0.116 |
| 106 |
248395_at |
AT5G52120
|
AtPP2-A14, phloem protein 2-A14, PP2-A14, phloem protein 2-A14 |
0.596 |
0.110 |
| 107 |
263653_at |
AT1G04310
|
ERS2, ethylene response sensor 2 |
0.596 |
0.177 |
| 108 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
0.596 |
0.014 |
| 109 |
252387_at |
AT3G47800
|
[Galactose mutarotase-like superfamily protein] |
0.595 |
0.331 |
| 110 |
253036_at |
AT4G38340
|
NLP3, NIN-like protein 3 |
0.595 |
0.037 |
| 111 |
264905_at |
AT2G17430
|
ATMLO7, MLO7, MILDEW RESISTANCE LOCUS O 7, NTA, NORTIA |
0.594 |
0.074 |
| 112 |
256697_at |
AT3G20660
|
AtOCT4, organic cation/carnitine transporter4, OCT4, organic cation/carnitine transporter4 |
0.591 |
0.276 |
| 113 |
255252_at |
AT4G04990
|
unknown |
0.589 |
0.105 |
| 114 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.588 |
-0.012 |
| 115 |
249066_at |
AT5G43920
|
[transducin family protein / WD-40 repeat family protein] |
0.584 |
0.063 |
| 116 |
249869_at |
AT5G23050
|
AAE17, acyl-activating enzyme 17 |
0.582 |
0.114 |
| 117 |
264551_at |
AT1G09460
|
[Carbohydrate-binding X8 domain superfamily protein] |
0.582 |
0.159 |
| 118 |
248224_at |
AT5G53490
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.582 |
0.147 |
| 119 |
250766_at |
AT5G05550
|
[sequence-specific DNA binding transcription factors] |
0.579 |
0.387 |
| 120 |
245543_at |
AT4G15260
|
[UDP-Glycosyltransferase superfamily protein] |
0.578 |
0.623 |
| 121 |
249234_at |
AT5G42200
|
[RING/U-box superfamily protein] |
0.576 |
0.198 |
| 122 |
260774_at |
AT1G78290
|
SNRK2-8, SNF1-RELATED PROTEIN KINASE 2-8, SNRK2.8, SNF1-RELATED PROTEIN KINASE 2.8, SRK2C, SNF1-RELATED PROTEIN KINASE 2C |
0.575 |
0.216 |
| 123 |
262502_at |
AT1G21600
|
PTAC6, plastid transcriptionally active 6 |
0.573 |
0.102 |
| 124 |
253666_at |
AT4G30270
|
MERI-5, MERISTEM 5, MERI5B, meristem-5, SEN4, SENESCENCE 4, XTH24, xyloglucan endotransglucosylase/hydrolase 24 |
0.571 |
0.229 |
| 125 |
256965_at |
AT3G13450
|
DIN4, DARK INDUCIBLE 4 |
0.571 |
0.647 |
| 126 |
262916_at |
AT1G59700
|
ATGSTU16, glutathione S-transferase TAU 16, GSTU16, glutathione S-transferase TAU 16 |
0.569 |
0.290 |
| 127 |
251066_at |
AT5G01880
|
DAFL2, DAF-Like gene 2 |
0.568 |
0.149 |
| 128 |
261842_at |
AT1G16160
|
WAKL5, wall associated kinase-like 5 |
0.568 |
0.033 |
| 129 |
260748_at |
AT1G49210
|
[RING/U-box superfamily protein] |
0.567 |
0.134 |
| 130 |
265902_at |
AT2G25590
|
[Plant Tudor-like protein] |
0.565 |
0.190 |
| 131 |
247136_at |
AT5G66170
|
STR18, sulfurtransferase 18 |
0.565 |
0.183 |
| 132 |
264580_at |
AT1G05340
|
unknown |
0.562 |
0.146 |
| 133 |
252570_at |
AT3G45300
|
ATIVD, IVD, isovaleryl-CoA-dehydrogenase, IVDH, ISOVALERYL-COA-DEHYDROGENASE |
0.560 |
0.539 |
| 134 |
260837_at |
AT1G43670
|
AtcFBP, Arabidopsis thaliana cytosolic fructose-1,6-bisphosphatase, FBP, fructose-1,6-bisphosphatase, FINS1, FRUCTOSE INSENSITIVE 1 |
0.558 |
0.248 |
| 135 |
263118_at |
AT1G03090
|
MCCA |
0.557 |
0.520 |
| 136 |
255521_at |
AT4G02280
|
ATSUS3, SUS3, sucrose synthase 3 |
0.553 |
-0.091 |
| 137 |
246929_at |
AT5G25210
|
unknown |
0.552 |
0.433 |
| 138 |
258281_at |
AT3G26900
|
ATSKL1, Arabidopsis thaliana shikimate kinase-like 1, SKL1, shikimate kinase-like 1 |
0.551 |
0.098 |
| 139 |
252924_at |
AT4G39070
|
BBX20, B-box domain protein 20, BZS1, BZS1 |
0.549 |
0.337 |
| 140 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.546 |
0.317 |
| 141 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
0.545 |
0.315 |
| 142 |
261294_at |
AT1G48430
|
[Dihydroxyacetone kinase] |
0.544 |
0.216 |
| 143 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
0.542 |
0.645 |
| 144 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.541 |
-0.114 |
| 145 |
253320_at |
AT4G34020
|
AtDJ1C, DJ-1 homolog C, DJ-1c, DJ1C, DJ-1 homolog C |
0.539 |
0.401 |
| 146 |
262643_at |
AT1G62770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.539 |
0.107 |
| 147 |
266224_at |
AT2G28800
|
ALB3, ALBINO 3 |
0.539 |
0.048 |
| 148 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.539 |
0.163 |
| 149 |
261453_at |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
0.539 |
0.304 |
| 150 |
251996_at |
AT3G52840
|
BGAL2, beta-galactosidase 2 |
0.538 |
0.102 |
| 151 |
262162_at |
AT1G78020
|
unknown |
0.537 |
-0.264 |
| 152 |
260205_at |
AT1G70700
|
JAZ9, JASMONATE-ZIM-DOMAIN PROTEIN 9, TIFY7 |
0.535 |
0.163 |
| 153 |
255181_at |
AT4G08110
|
[CACTA-like transposase family (Ptta/En/Spm), has a 4.2e-66 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
0.534 |
-0.003 |
| 154 |
264843_at |
AT1G03400
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.534 |
0.302 |
| 155 |
261100_at |
AT1G63020
|
NRPD1, NRPD1A, nuclear RNA polymerase D1A, POL IVA, NUCLEAR RNA POLYMERASE D 1A, SDE4, SMD2, SILENCING MOVEMENT DEFICIENT 2 |
0.533 |
0.034 |
| 156 |
245659_at |
AT1G28260
|
[Telomerase activating protein Est1] |
0.531 |
0.475 |
| 157 |
256499_at |
AT1G36640
|
unknown |
0.531 |
0.077 |
| 158 |
253160_at |
AT4G35760
|
LTO1, Lumen Thiol Oxidoreductase 1 |
0.530 |
0.090 |
| 159 |
260966_at |
AT1G12220
|
RPS5, RESISTANT TO P. SYRINGAE 5 |
0.529 |
0.217 |
| 160 |
264700_at |
AT1G70100
|
unknown |
0.529 |
0.319 |
| 161 |
265189_at |
AT1G23840
|
unknown |
0.527 |
0.184 |
| 162 |
266695_at |
AT2G19810
|
AtOZF1, AtTZF2, OZF1, Oxidation-related Zinc Finger 1, TZF2, tandem zinc finger 2 |
0.527 |
0.263 |
| 163 |
260272_at |
AT1G80570
|
[RNI-like superfamily protein] |
0.526 |
0.281 |
| 164 |
247525_at |
AT5G61380
|
APRR1, AtTOC1, TIMING OF CAB EXPRESSION 1, PRR1, PSEUDO-RESPONSE REGULATOR 1, TOC1, TIMING OF CAB EXPRESSION 1 |
0.526 |
0.164 |
| 165 |
255602_at |
AT4G01026
|
PYL7, PYR1-like 7, RCAR2, regulatory components of ABA receptor 2 |
0.524 |
0.369 |
| 166 |
247617_at |
AT5G60270
|
LecRK-I.7, L-type lectin receptor kinase I.7 |
0.524 |
0.123 |
| 167 |
248029_at |
AT5G55700
|
BAM4, beta-amylase 4, BMY6, BETA-AMYLASE 6 |
0.524 |
0.588 |
| 168 |
253829_at |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
0.523 |
0.477 |
| 169 |
253279_at |
AT4G34030
|
MCCB, 3-methylcrotonyl-CoA carboxylase |
0.518 |
0.449 |
| 170 |
258603_at |
AT3G02990
|
ATHSFA1E, heat shock transcription factor A1E, HSFA1E, heat shock transcription factor A1E |
0.517 |
0.185 |
| 171 |
249824_at |
AT5G23380
|
unknown |
0.514 |
0.342 |
| 172 |
251356_at |
AT3G61060
|
AtPP2-A13, phloem protein 2-A13, PP2-A13, phloem protein 2-A13 |
0.513 |
0.359 |
| 173 |
251245_at |
AT3G62090
|
PIF6, PHYTOCHROME-INTERACTING FACTOR 6, PIL2, phytochrome interacting factor 3-like 2 |
0.511 |
-0.006 |
| 174 |
248193_at |
AT5G54080
|
AtHGO, HGO, homogentisate 1,2-dioxygenase |
0.510 |
0.532 |
| 175 |
246703_at |
AT5G28080
|
WNK9 |
0.510 |
0.308 |
| 176 |
257772_at |
AT3G23080
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.510 |
0.350 |
| 177 |
246493_at |
AT5G16180
|
ATCRS1, ARABIDOPSIS ORTHOLOG OF MAIZE CHLOROPLAST SPLICING FACTOR CRS1, CRS1, ortholog of maize chloroplast splicing factor CRS1 |
0.510 |
0.270 |
| 178 |
264199_at |
AT1G22700
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.509 |
0.064 |
| 179 |
249415_at |
AT5G39660
|
CDF2, cycling DOF factor 2 |
0.509 |
0.021 |
| 180 |
264405_at |
AT1G10210
|
ATMPK1, mitogen-activated protein kinase 1, MPK1, mitogen-activated protein kinase 1 |
0.508 |
0.225 |
| 181 |
263196_at |
AT1G36070
|
[Transducin/WD40 repeat-like superfamily protein] |
0.508 |
0.397 |
| 182 |
259008_at |
AT3G09390
|
ATMT-1, ARABIDOPSIS THALIANA METALLOTHIONEIN-1, ATMT-K, ARABIDOPSIS THALIANA METALLOTHIONEIN-K, MT2A, metallothionein 2A |
0.505 |
0.032 |
| 183 |
267035_at |
AT2G38400
|
AGT3, alanine:glyoxylate aminotransferase 3 |
0.504 |
0.586 |
| 184 |
252315_at |
AT3G48690
|
ATCXE12, ARABIDOPSIS THALIANA CARBOXYESTERASE 12, CXE12 |
0.504 |
0.178 |
| 185 |
265192_at |
AT1G05060
|
unknown |
0.504 |
0.098 |
| 186 |
245925_at |
AT5G28770
|
AtbZIP63, Arabidopsis thaliana basic leucine zipper 63, BZO2H3 |
0.503 |
0.082 |
| 187 |
253776_at |
AT4G28390
|
AAC3, ADP/ATP carrier 3, ATAAC3 |
0.502 |
0.104 |
| 188 |
251719_at |
AT3G56140
|
unknown |
0.501 |
0.217 |
| 189 |
254384_at |
AT4G21870
|
[HSP20-like chaperones superfamily protein] |
0.499 |
-0.153 |
| 190 |
262452_at |
AT1G11210
|
unknown |
0.497 |
0.126 |
| 191 |
251603_at |
AT3G57760
|
[Protein kinase superfamily protein] |
0.494 |
0.403 |
| 192 |
248148_at |
AT5G54930
|
[AT hook motif-containing protein] |
0.493 |
0.063 |
| 193 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
0.491 |
0.707 |
| 194 |
264236_at |
AT1G54680
|
unknown |
0.490 |
0.391 |
| 195 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
0.487 |
0.310 |
| 196 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
0.485 |
0.253 |
| 197 |
250752_at |
AT5G05690
|
CBB3, CABBAGE 3, CPD, CONSTITUTIVE PHOTOMORPHOGENIC DWARF, CYP90, CYP90A, CYP90A1, CYTOCHROME P450 90A1, DWF3, DWARF 3 |
0.483 |
0.325 |
| 198 |
245198_at |
AT1G67700
|
HHL1, HYPERSENSITIVE TO HIGH LIGHT 1 |
0.483 |
0.045 |
| 199 |
255540_at |
AT4G01800
|
AGY1, Albino or Glassy Yellow 1, AtcpSecA, Arabidopsis thaliana chloroplast SecA, SECA1 |
0.482 |
0.123 |
| 200 |
250248_at |
AT5G13740
|
ZIF1, zinc induced facilitator 1 |
0.482 |
-0.137 |
| 201 |
251100_at |
AT5G01670
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.479 |
0.280 |
| 202 |
265330_at |
AT2G18440
|
GUT15, GENE WITH UNSTABLE TRANSCRIPT 15 |
0.479 |
0.027 |
| 203 |
247774_at |
AT5G58660
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.479 |
0.155 |
| 204 |
256061_at |
AT1G07040
|
unknown |
0.476 |
0.514 |
| 205 |
254105_at |
AT4G25080
|
CHLM, magnesium-protoporphyrin IX methyltransferase |
0.476 |
0.021 |
| 206 |
247278_at |
AT5G64380
|
[Inositol monophosphatase family protein] |
0.476 |
0.267 |
| 207 |
256796_at |
AT3G22210
|
unknown |
0.474 |
-0.210 |
| 208 |
255436_at |
AT4G03150
|
unknown |
0.474 |
-0.174 |
| 209 |
247532_at |
AT5G61560
|
[U-box domain-containing protein kinase family protein] |
0.472 |
0.104 |
| 210 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
0.472 |
0.516 |
| 211 |
251884_at |
AT3G54150
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.471 |
0.218 |
| 212 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
0.471 |
0.219 |
| 213 |
266583_at |
AT2G46220
|
[Uncharacterized conserved protein (DUF2358)] |
0.470 |
0.163 |
| 214 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
0.470 |
0.033 |
| 215 |
254206_at |
AT4G24180
|
ATTLP1, TLP1, THAUMATIN-LIKE PROTEIN 1 |
0.469 |
0.073 |
| 216 |
250710_at |
AT5G06100
|
ATMYB33, MYB33, myb domain protein 33 |
0.468 |
0.058 |
| 217 |
261507_at |
AT1G71720
|
PDE338, PIGMENT DEFECTIVE 338, RLSB, RbcL mRNA S1 binding domain protein |
0.464 |
0.063 |
| 218 |
254787_at |
AT4G12690
|
unknown |
0.463 |
0.331 |
| 219 |
258100_at |
AT3G23550
|
[MATE efflux family protein] |
0.462 |
0.130 |
| 220 |
246028_at |
AT5G21170
|
5'-AMP-activated protein kinase beta-2 subunit protein |
0.462 |
0.377 |
| 221 |
264082_at |
AT2G28570
|
unknown |
0.460 |
0.058 |
| 222 |
260465_at |
AT1G10910
|
EMB3103, EMBRYO DEFECTIVE 3103 |
0.460 |
0.040 |
| 223 |
260760_at |
AT1G49170
|
unknown |
0.459 |
0.371 |
| 224 |
259517_at |
AT1G20630
|
CAT1, catalase 1 |
0.459 |
0.374 |
| 225 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
0.457 |
0.178 |
| 226 |
259058_at |
AT3G03470
|
CYP89A9, cytochrome P450, family 87, subfamily A, polypeptide 9 |
0.455 |
0.427 |
| 227 |
251022_at |
AT5G02150
|
Fes1C, Fes1C |
0.455 |
0.488 |
| 228 |
266839_at |
AT2G25930
|
ELF3, EARLY FLOWERING 3, PYK20 |
0.454 |
0.215 |
| 229 |
263708_at |
AT1G09320
|
[agenet domain-containing protein] |
0.454 |
0.284 |
| 230 |
254797_at |
AT4G13030
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.454 |
0.287 |
| 231 |
254669_at |
AT4G18370
|
DEG5, degradation of periplasmic proteins 5, DEGP5, DEGP PROTEASE 5, HHOA, PROTEASE HHOA PRECUSOR |
0.454 |
0.071 |
| 232 |
249393_at |
AT5G40170
|
AtRLP54, receptor like protein 54, RLP54, receptor like protein 54 |
0.454 |
-0.045 |
| 233 |
252979_at |
AT4G38225
|
unknown |
0.453 |
0.131 |
| 234 |
266335_at |
AT2G32440
|
ATKAO2, ARABIDOPSIS ENT-KAURENOIC ACID HYDROXYLASE 2, CYP88A4, KAO2, ent-kaurenoic acid hydroxylase 2 |
0.453 |
0.135 |
| 235 |
262505_at |
AT1G21680
|
[DPP6 N-terminal domain-like protein] |
0.453 |
0.436 |
| 236 |
247540_at |
AT5G61590
|
[Integrase-type DNA-binding superfamily protein] |
0.452 |
0.128 |
| 237 |
254545_at |
AT4G19830
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
0.452 |
-0.020 |
| 238 |
248901_at |
AT5G46410
|
SSP4, SCP1-like small phosphatase 4 |
0.451 |
0.286 |
| 239 |
247079_at |
AT5G66055
|
AKRP, ankyrin repeat protein, EMB16, EMBRYO DEFECTIVE 16, EMB2036, EMBRYO DEFECTIVE 2036 |
0.451 |
0.118 |
| 240 |
246393_at |
AT1G58150
|
unknown |
0.451 |
-0.008 |
| 241 |
260361_at |
AT1G69360
|
unknown |
0.450 |
0.226 |
| 242 |
263836_at |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
0.450 |
-0.134 |
| 243 |
245418_at |
AT4G17370
|
[Oxidoreductase family protein] |
0.450 |
0.046 |
| 244 |
249929_at |
AT5G22340
|
unknown |
0.449 |
0.058 |
| 245 |
255452_at |
AT4G02880
|
unknown |
0.448 |
0.323 |
| 246 |
255285_at |
AT4G04630
|
unknown |
0.447 |
0.422 |
| 247 |
254409_at |
AT4G21400
|
CRK28, cysteine-rich RLK (RECEPTOR-like protein kinase) 28 |
0.446 |
0.012 |
| 248 |
250773_at |
AT5G05430
|
[RNA-binding protein] |
0.445 |
-0.030 |
| 249 |
263593_at |
AT2G01860
|
EMB975, EMBRYO DEFECTIVE 975 |
0.445 |
0.383 |
| 250 |
250913_at |
AT5G03770
|
AtKdtA, KDTA, KDO transferase A |
0.445 |
0.228 |
| 251 |
264954_at |
AT1G77060
|
[Phosphoenolpyruvate carboxylase family protein] |
0.444 |
0.250 |
| 252 |
250776_at |
AT5G05320
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
0.444 |
0.197 |
| 253 |
247130_at |
AT5G66180
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.444 |
0.291 |
| 254 |
266018_at |
AT2G18710
|
SCY1, SECY homolog 1 |
0.443 |
0.095 |
| 255 |
248991_at |
AT5G45220
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.442 |
0.090 |
| 256 |
262539_at |
AT1G17200
|
[Uncharacterised protein family (UPF0497)] |
0.442 |
0.001 |
| 257 |
252990_at |
AT4G38440
|
IYO, MINIYO |
0.442 |
0.072 |
| 258 |
260451_at |
AT1G72360
|
AtERF73, ERF73, ethylene response factor 73, HRE1, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 1 |
0.441 |
0.340 |
| 259 |
251414_at |
AT3G60370
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
0.441 |
0.071 |
| 260 |
263909_at |
AT2G36490
|
AtROS1, DML1, demeter-like 1, ROS1, REPRESSOR OF SILENCING1 |
0.441 |
0.124 |
| 261 |
257990_at |
AT3G19860
|
bHLH121, basic Helix-Loop-Helix 121 |
0.441 |
0.247 |
| 262 |
266917_at |
AT2G45830
|
DTA2, downstream target of AGL15 2 |
0.440 |
-0.024 |
| 263 |
247123_at |
AT5G66050
|
[Wound-responsive family protein] |
0.440 |
0.281 |
| 264 |
249010_at |
AT5G44580
|
unknown |
0.440 |
-0.327 |
| 265 |
259340_at |
AT3G03870
|
unknown |
0.440 |
0.158 |
| 266 |
261596_at |
AT1G33080
|
[MATE efflux family protein] |
0.439 |
0.193 |
| 267 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
0.439 |
0.002 |
| 268 |
249389_at |
AT5G40100
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.439 |
0.192 |
| 269 |
267379_at |
AT2G26340
|
unknown |
0.439 |
0.069 |
| 270 |
256071_at |
AT1G13640
|
[Phosphatidylinositol 3- and 4-kinase family protein] |
0.438 |
0.042 |
| 271 |
253108_at |
AT4G35900
|
atbzip14, FD, FD-1 |
0.438 |
0.057 |
| 272 |
254032_at |
AT4G25940
|
[ENTH/ANTH/VHS superfamily protein] |
0.437 |
0.106 |
| 273 |
254681_at |
AT4G18140
|
SSP4b, SCP1-like small phosphatase 4b |
0.437 |
0.264 |
| 274 |
263647_at |
AT2G04690
|
Pyridoxamine 5'-phosphate oxidase family protein |
0.436 |
0.356 |
| 275 |
248432_at |
AT5G51390
|
unknown |
0.436 |
0.572 |
| 276 |
254424_at |
AT4G21510
|
AtFBS2, FBS2, F-box stress induced 2 |
0.436 |
0.033 |
| 277 |
251133_at |
AT5G01240
|
LAX1, like AUXIN RESISTANT 1 |
0.435 |
0.186 |
| 278 |
249839_at |
AT5G23405
|
[HMG-box (high mobility group) DNA-binding family protein] |
0.435 |
0.216 |
| 279 |
253373_at |
AT4G33150
|
LKR, LKR/SDH, LYSINE-KETOGLUTARATE REDUCTASE/SACCHAROPINE DEHYDROGENASE, SDH, SACCHAROPINE DEHYDROGENASE |
0.435 |
0.464 |
| 280 |
253747_at |
AT4G29050
|
LecRK-V.9, L-type lectin receptor kinase V.9 |
0.435 |
0.129 |
| 281 |
254919_at |
AT4G11360
|
RHA1B, RING-H2 finger A1B |
0.435 |
0.087 |
| 282 |
247616_at |
AT5G60260
|
unknown |
0.434 |
0.048 |
| 283 |
256394_at |
AT3G06290
|
AtSAC3B, SAC3B, yeast Sac3 homolog B |
0.434 |
0.147 |
| 284 |
246875_at |
AT5G26130
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.434 |
0.093 |
| 285 |
255148_at |
AT4G08470
|
MAPKKK10, MEKK3, MAPK/ERK kinase kinase 3 |
0.433 |
0.220 |
| 286 |
245807_at |
AT1G46768
|
RAP2.1, related to AP2 1 |
0.432 |
0.164 |
| 287 |
260936_at |
AT1G45150
|
unknown |
0.432 |
0.210 |
| 288 |
261272_at |
AT1G26665
|
[Mediator complex, subunit Med10] |
0.432 |
0.309 |
| 289 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
0.431 |
0.168 |
| 290 |
247710_at |
AT5G59260
|
LecRK-II.1, L-type lectin receptor kinase II.1 |
0.431 |
0.211 |
| 291 |
248166_at |
AT5G54520
|
[Transducin/WD40 repeat-like superfamily protein] |
0.430 |
0.304 |
| 292 |
249101_at |
AT5G43580
|
UPI, UNUSUAL SERINE PROTEASE INHIBITOR |
0.430 |
-0.009 |
| 293 |
263365_at |
AT2G20550
|
[HSP40/DnaJ peptide-binding protein] |
0.429 |
0.097 |
| 294 |
259118_at |
AT3G01310
|
[Phosphoglycerate mutase-like family protein] |
0.429 |
0.348 |
| 295 |
249750_at |
AT5G24570
|
unknown |
0.428 |
-0.142 |
| 296 |
260759_at |
AT1G49180
|
[protein kinase family protein] |
0.428 |
0.122 |
| 297 |
253624_at |
AT4G30580
|
ATS2, EMB1995, EMBRYO DEFECTIVE 1995, LPAT1, lysophosphatidic acid acyltransferase 1 |
0.428 |
0.097 |
| 298 |
250017_at |
AT5G18140
|
DJC69, DNA J protein A69 |
0.428 |
0.196 |
| 299 |
248564_at |
AT5G49700
|
AHL17, AT-hook motif nuclear localized protein 17 |
0.427 |
-0.016 |
| 300 |
250938_at |
AT5G03180
|
[RING/U-box superfamily protein] |
0.425 |
0.170 |
| 301 |
253502_at |
AT4G31940
|
CYP82C4, cytochrome P450, family 82, subfamily C, polypeptide 4 |
-1.608 |
-0.841 |
| 302 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
-1.486 |
-1.200 |
| 303 |
246652_at |
AT5G35190
|
EXT13, extensin 13 |
-1.318 |
-0.453 |
| 304 |
254907_at |
AT4G11190
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-1.301 |
-0.538 |
| 305 |
259388_at |
AT1G13420
|
ATST4B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 4B, ST4B, sulfotransferase 4B |
-1.221 |
-0.249 |
| 306 |
260745_at |
AT1G78370
|
ATGSTU20, glutathione S-transferase TAU 20, GSTU20, glutathione S-transferase TAU 20 |
-1.209 |
-0.597 |
| 307 |
254056_at |
AT4G25250
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-1.204 |
-0.307 |
| 308 |
262373_at |
AT1G73120
|
unknown |
-1.166 |
0.063 |
| 309 |
257135_at |
AT3G12900
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-1.150 |
-0.365 |
| 310 |
253040_at |
AT4G37800
|
XTH7, xyloglucan endotransglucosylase/hydrolase 7 |
-1.142 |
-0.377 |
| 311 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
-1.130 |
-0.116 |
| 312 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
-1.123 |
-0.284 |
| 313 |
246229_at |
AT4G37160
|
sks15, SKU5 similar 15 |
-1.109 |
-0.678 |
| 314 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
-1.076 |
-0.730 |
| 315 |
254909_at |
AT4G11210
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-1.061 |
-0.338 |
| 316 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
-1.056 |
-0.247 |
| 317 |
260067_at |
AT1G73780
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-1.054 |
-0.054 |
| 318 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
-1.027 |
-0.696 |
| 319 |
253684_at |
AT4G29690
|
[Alkaline-phosphatase-like family protein] |
-1.010 |
-0.576 |
| 320 |
247871_at |
AT5G57530
|
AtXTH12, XTH12, xyloglucan endotransglucosylase/hydrolase 12 |
-1.001 |
-0.187 |
| 321 |
247483_at |
AT5G62420
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.990 |
-0.201 |
| 322 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.971 |
-0.719 |
| 323 |
246872_at |
AT5G26080
|
[proline-rich family protein] |
-0.966 |
-0.081 |
| 324 |
259681_at |
AT1G77760
|
GNR1, NIA1, nitrate reductase 1, NR1, NITRATE REDUCTASE 1 |
-0.958 |
-0.099 |
| 325 |
267222_at |
AT2G43880
|
[Pectin lyase-like superfamily protein] |
-0.955 |
-0.278 |
| 326 |
248048_at |
AT5G56080
|
ATNAS2, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 2, NAS2, nicotianamine synthase 2 |
-0.953 |
-0.318 |
| 327 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
-0.941 |
-0.176 |
| 328 |
250661_at |
AT5G07030
|
[Eukaryotic aspartyl protease family protein] |
-0.930 |
-0.409 |
| 329 |
247070_at |
AT5G66815
|
unknown |
-0.923 |
-0.103 |
| 330 |
245967_at |
AT5G19800
|
[hydroxyproline-rich glycoprotein family protein] |
-0.913 |
-0.130 |
| 331 |
251231_at |
AT3G62760
|
ATGSTF13 |
-0.911 |
-0.170 |
| 332 |
264672_at |
AT1G09750
|
[Eukaryotic aspartyl protease family protein] |
-0.900 |
0.155 |
| 333 |
257203_at |
AT3G23730
|
XTH16, xyloglucan endotransglucosylase/hydrolase 16 |
-0.893 |
-0.186 |
| 334 |
252232_at |
AT3G49760
|
AtbZIP5, basic leucine-zipper 5, bZIP5, basic leucine-zipper 5 |
-0.888 |
-0.147 |
| 335 |
248727_at |
AT5G47990
|
CYP705A5, cytochrome P450, family 705, subfamily A, polypeptide 5, THAD, THALIAN-DIOL DESATURASE, THAD1, THALIAN-DIOL DESATURASE |
-0.885 |
-0.448 |
| 336 |
249599_at |
AT5G37990
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.883 |
-0.771 |
| 337 |
250801_at |
AT5G04960
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.879 |
-0.431 |
| 338 |
249756_at |
AT5G24313
|
unknown |
-0.877 |
0.012 |
| 339 |
262978_at |
AT1G75780
|
TUB1, tubulin beta-1 chain |
-0.875 |
-0.235 |
| 340 |
253309_at |
AT4G33790
|
CER4, ECERIFERUM 4, FAR3, FATTY ACID REDUCTASE 3, G7 |
-0.873 |
-0.434 |
| 341 |
248684_at |
AT5G48485
|
DIR1, DEFECTIVE IN INDUCED RESISTANCE 1 |
-0.872 |
0.056 |
| 342 |
264404_at |
AT2G25160
|
CYP82F1, cytochrome P450, family 82, subfamily F, polypeptide 1 |
-0.870 |
-0.299 |
| 343 |
249522_at |
AT5G38700
|
unknown |
-0.869 |
0.040 |
| 344 |
258008_at |
AT3G19430
|
[late embryogenesis abundant protein-related / LEA protein-related] |
-0.869 |
-0.379 |
| 345 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
-0.865 |
-0.432 |
| 346 |
248728_at |
AT5G48000
|
CYP708 A2, CYTOCHROME P450, FAMILY 708, SUBFAMILY A, POLYPEPTIDE 2, CYP708A2, cytochrome P450, family 708, subfamily A, polypeptide 2, THAH, THALIANOL HYDROXYLASE, THAH1, THALIANOL HYDROXYLASE 1 |
-0.863 |
-0.609 |
| 347 |
262198_at |
AT1G53830
|
ATPME2, pectin methylesterase 2, PME2, pectin methylesterase 2 |
-0.862 |
-0.346 |
| 348 |
259064_at |
AT3G07490
|
AGD11, ARF-GAP domain 11, AtCML3, CML3, calmodulin-like 3 |
-0.860 |
-0.056 |
| 349 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
-0.856 |
-0.323 |
| 350 |
267287_at |
AT2G23630
|
sks16, SKU5 similar 16 |
-0.853 |
-0.311 |
| 351 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
-0.847 |
-0.126 |
| 352 |
267101_at |
AT2G41480
|
AtPRX25, PRX25, peroxidase 25 |
-0.846 |
-0.384 |
| 353 |
251952_at |
AT3G53650
|
[Histone superfamily protein] |
-0.844 |
0.025 |
| 354 |
247914_at |
AT5G57540
|
AtXTH13, XTH13, xyloglucan endotransglucosylase/hydrolase 13 |
-0.839 |
-0.069 |
| 355 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
-0.833 |
-0.285 |
| 356 |
250892_at |
AT5G03760
|
ATCSLA09, ATCSLA9, CSLA09, CSLA9, CELLULOSE SYNTHASE LIKE A9, RAT4, RESISTANT TO AGROBACTERIUM TRANSFORMATION 4 |
-0.832 |
-0.583 |
| 357 |
265083_at |
AT1G03820
|
unknown |
-0.831 |
-0.447 |
| 358 |
249203_at |
AT5G42590
|
CYP71A16, cytochrome P450, family 71, subfamily A, polypeptide 16, MRO, marneral oxidase |
-0.825 |
-0.170 |
| 359 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.823 |
-1.116 |
| 360 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
-0.817 |
-0.291 |
| 361 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.816 |
-0.293 |
| 362 |
264640_at |
AT1G65680
|
ATEXPB2, expansin B2, ATHEXP BETA 1.4, EXPB2, expansin B2 |
-0.816 |
-0.068 |
| 363 |
245689_at |
AT5G04120
|
[Phosphoglycerate mutase family protein] |
-0.811 |
-0.448 |
| 364 |
254534_at |
AT4G19680
|
ATIRT2, IRON REGULATED TRANSPORTER 2, IRT2, iron regulated transporter 2 |
-0.807 |
-0.401 |
| 365 |
264403_at |
AT2G25150
|
[HXXXD-type acyl-transferase family protein] |
-0.807 |
-0.089 |
| 366 |
253763_at |
AT4G28850
|
ATXTH26, XTH26, xyloglucan endotransglucosylase/hydrolase 26 |
-0.805 |
0.023 |
| 367 |
253949_at |
AT4G26780
|
AR192, MGE2, mitochondrial GrpE 2 |
-0.802 |
-0.698 |
| 368 |
266191_at |
AT2G39040
|
[Peroxidase superfamily protein] |
-0.799 |
-0.291 |
| 369 |
245904_at |
AT5G11110
|
ATSPS2F, sucrose phosphate synthase 2F, KNS2, KAONASHI 2, SPS1, SUCROSE PHOSPHATE SYNTHASE 1, SPS2F, sucrose phosphate synthase 2F, SPSA2, sucrose-phosphate synthase A2 |
-0.793 |
-0.327 |
| 370 |
257645_at |
AT3G25790
|
[myb-like transcription factor family protein] |
-0.791 |
-0.429 |
| 371 |
248729_at |
AT5G48010
|
AtTHAS1, Arabidopsis thaliana thalianol synthase 1, THAS, THALIANOL SYNTHASE, THAS1, thalianol synthase 1 |
-0.789 |
-0.527 |
| 372 |
252858_at |
AT4G39770
|
TPPH, trehalose-6-phosphate phosphatase H |
-0.784 |
-0.170 |
| 373 |
260077_at |
AT1G73620
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.782 |
-0.467 |
| 374 |
249567_at |
AT5G38020
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.781 |
-0.489 |
| 375 |
247097_at |
AT5G66460
|
AtMAN7, MAN7, endo-beta-mannase 7 |
-0.778 |
-0.340 |
| 376 |
255450_at |
AT4G02850
|
[phenazine biosynthesis PhzC/PhzF family protein] |
-0.777 |
-0.179 |
| 377 |
258765_at |
AT3G10710
|
RHS12, root hair specific 12 |
-0.776 |
-0.308 |
| 378 |
254550_at |
AT4G19690
|
ATIRT1, ARABIDOPSIS IRON-REGULATED TRANSPORTER 1, IRT1, iron-regulated transporter 1 |
-0.775 |
-1.090 |
| 379 |
253598_at |
AT4G30800
|
[Nucleic acid-binding, OB-fold-like protein] |
-0.772 |
-0.579 |
| 380 |
248963_at |
AT5G45700
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.771 |
-0.191 |
| 381 |
261574_at |
AT1G01190
|
CYP78A8, cytochrome P450, family 78, subfamily A, polypeptide 8 |
-0.769 |
-0.361 |
| 382 |
264347_at |
AT1G12040
|
LRX1, leucine-rich repeat/extensin 1 |
-0.768 |
-0.233 |
| 383 |
248178_at |
AT5G54370
|
[Late embryogenesis abundant (LEA) protein-related] |
-0.767 |
-0.180 |
| 384 |
249063_at |
AT5G44110
|
ABCI21, ATP-binding cassette A21, ATNAP2, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN, ATPOP1, POP1 |
-0.767 |
-0.375 |
| 385 |
265766_at |
AT2G48080
|
[oxidoreductase, 2OG-Fe(II) oxygenase family protein] |
-0.765 |
-0.036 |
| 386 |
262838_at |
AT1G14960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.764 |
-0.308 |
| 387 |
246827_at |
AT5G26330
|
[Cupredoxin superfamily protein] |
-0.763 |
-0.175 |
| 388 |
247268_at |
AT5G64080
|
AtXYP1, XYP1, xylogen protein 1 |
-0.753 |
-0.145 |
| 389 |
246403_at |
AT1G57590
|
[Pectinacetylesterase family protein] |
-0.753 |
-0.303 |
| 390 |
246395_at |
AT1G58170
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.752 |
-0.182 |
| 391 |
254107_at |
AT4G25220
|
AtG3Pp2, glycerol-3-phosphate permease 2, G3Pp2, glycerol-3-phosphate permease 2, RHS15, root hair specific 15 |
-0.750 |
0.095 |
| 392 |
263594_at |
AT2G01880
|
ATPAP7, PURPLE ACID PHOSPHATASE 7, PAP7, purple acid phosphatase 7 |
-0.747 |
-0.232 |
| 393 |
253515_at |
AT4G31320
|
SAUR37, SMALL AUXIN UPREGULATED RNA 37 |
-0.744 |
-0.422 |
| 394 |
250469_at |
AT5G10130
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.741 |
-0.349 |
| 395 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.739 |
-0.333 |
| 396 |
248732_at |
AT5G48070
|
ATXTH20, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 20, XTH20, xyloglucan endotransglucosylase/hydrolase 20 |
-0.735 |
-0.317 |
| 397 |
248725_at |
AT5G47980
|
[HXXXD-type acyl-transferase family protein] |
-0.734 |
-0.161 |
| 398 |
253088_at |
AT4G36220
|
CYP84A1, CYTOCHROME P450 84A1, FAH1, ferulic acid 5-hydroxylase 1 |
-0.730 |
-0.470 |
| 399 |
246920_at |
AT5G25090
|
AtENODL13, ENODL13, early nodulin-like protein 13 |
-0.729 |
-0.550 |
| 400 |
247706_at |
AT5G59480
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.723 |
-0.215 |
| 401 |
266765_at |
AT2G46860
|
AtPPa3, pyrophosphorylase 3, PPa3, pyrophosphorylase 3 |
-0.723 |
-0.107 |
| 402 |
258618_at |
AT3G02885
|
GASA5, GAST1 protein homolog 5 |
-0.722 |
-0.015 |
| 403 |
253832_at |
AT4G27654
|
unknown |
-0.718 |
0.427 |
| 404 |
266828_at |
AT2G22930
|
[UDP-Glycosyltransferase superfamily protein] |
-0.716 |
-0.126 |
| 405 |
266352_at |
AT2G01610
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.713 |
-0.142 |
| 406 |
245966_at |
AT5G19790
|
RAP2.11, related to AP2 11 |
-0.711 |
-0.176 |
| 407 |
254025_at |
AT4G25790
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.711 |
-0.223 |
| 408 |
248408_at |
AT5G51520
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.709 |
-0.106 |
| 409 |
261099_at |
AT1G62980
|
ATEXP18, ATEXPA18, expansin A18, ATHEXP ALPHA 1.25, EXP18, EXPANSIN 18, EXPA18, expansin A18 |
-0.709 |
-0.361 |
| 410 |
263553_at |
AT2G16430
|
ATPAP10, PAP10, purple acid phosphatase 10 |
-0.707 |
-0.282 |
| 411 |
249934_at |
AT5G22410
|
RHS18, root hair specific 18 |
-0.706 |
-0.022 |
| 412 |
245318_at |
AT4G16980
|
[arabinogalactan-protein family] |
-0.706 |
-0.137 |
| 413 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
-0.704 |
-0.000 |
| 414 |
252202_at |
AT3G50300
|
[HXXXD-type acyl-transferase family protein] |
-0.703 |
-0.326 |
| 415 |
245674_at |
AT1G56680
|
[Chitinase family protein] |
-0.702 |
-0.067 |
| 416 |
254837_at |
AT4G12360
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.701 |
0.031 |
| 417 |
263677_at |
AT1G04520
|
PDLP2, plasmodesmata-located protein 2 |
-0.701 |
-0.352 |
| 418 |
253286_at |
AT4G34260
|
AXY8, ALTERED XYLOGLUCAN 8, FUC95A |
-0.698 |
-0.060 |
| 419 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
-0.696 |
-0.259 |
| 420 |
250651_at |
AT5G06900
|
CYP93D1, cytochrome P450, family 93, subfamily D, polypeptide 1 |
-0.696 |
0.025 |
| 421 |
256779_at |
AT3G13784
|
AtcwINV5, cell wall invertase 5, CWINV5, cell wall invertase 5 |
-0.694 |
0.100 |
| 422 |
258013_at |
AT3G19320
|
[Leucine-rich repeat (LRR) family protein] |
-0.694 |
-0.126 |
| 423 |
257720_at |
AT3G18450
|
[PLAC8 family protein] |
-0.693 |
-0.021 |
| 424 |
256125_at |
AT1G18250
|
ATLP-1 |
-0.693 |
-0.465 |
| 425 |
265463_at |
AT2G37090
|
IRX9, IRREGULAR XYLEM 9 |
-0.691 |
-0.175 |
| 426 |
262704_at |
AT1G16530
|
ASL9, ASYMMETRIC LEAVES 2-like 9, LBD3, LOB DOMAIN-CONTAINING PROTEIN 3 |
-0.690 |
-0.056 |
| 427 |
255773_at |
AT1G18590
|
ATSOT17, SULFOTRANSFERASE 17, ATST5C, ARABIDOPSIS SULFOTRANSFERASE 5C, SOT17, sulfotransferase 17 |
-0.688 |
-0.653 |
| 428 |
255814_at |
AT1G19900
|
[glyoxal oxidase-related protein] |
-0.687 |
-0.288 |
| 429 |
249971_at |
AT5G19110
|
[Eukaryotic aspartyl protease family protein] |
-0.687 |
-0.057 |
| 430 |
263942_at |
AT2G35860
|
FLA16, FASCICLIN-like arabinogalactan protein 16 precursor |
-0.686 |
-0.546 |
| 431 |
251668_at |
AT3G57010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.685 |
-0.543 |
| 432 |
260119_at |
AT1G33930
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.684 |
0.091 |
| 433 |
257735_at |
AT3G27400
|
[Pectin lyase-like superfamily protein] |
-0.678 |
-0.066 |
| 434 |
258859_at |
AT3G02120
|
[hydroxyproline-rich glycoprotein family protein] |
-0.677 |
-0.218 |
| 435 |
266965_at |
AT2G39510
|
UMAMIT14, Usually multiple acids move in and out Transporters 14 |
-0.676 |
-0.358 |
| 436 |
252070_at |
AT3G51680
|
AtSDR2, SDR2, short-chain dehydrogenase/reductase 2 |
-0.674 |
-0.170 |
| 437 |
259195_at |
AT3G01730
|
unknown |
-0.673 |
0.021 |
| 438 |
255818_at |
AT2G33570
|
GALS1, galactan synthase 1 |
-0.672 |
0.076 |
| 439 |
266253_at |
AT2G27840
|
HD2D, histone deacetylase 2D, HDA13, HISTONE DEACETYLASE 13, HDT04, HDT4 |
-0.672 |
-0.640 |
| 440 |
266790_at |
AT2G28950
|
ATEXP6, ARABIDOPSIS THALIANA TEXPANSIN 6, ATEXPA6, expansin A6, ATHEXP ALPHA 1.8, EXPA6, expansin A6 |
-0.669 |
-0.248 |
| 441 |
254119_at |
AT4G24780
|
[Pectin lyase-like superfamily protein] |
-0.665 |
-0.430 |
| 442 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
-0.664 |
-0.397 |
| 443 |
257774_at |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
-0.664 |
-0.257 |
| 444 |
247214_at |
AT5G64850
|
unknown |
-0.664 |
-0.410 |
| 445 |
254632_at |
AT4G18630
|
unknown |
-0.663 |
-0.061 |
| 446 |
251287_at |
AT3G61820
|
[Eukaryotic aspartyl protease family protein] |
-0.663 |
-0.364 |
| 447 |
256600_at |
AT3G14850
|
TBL41, TRICHOME BIREFRINGENCE-LIKE 41 |
-0.662 |
-0.206 |
| 448 |
255284_at |
AT4G04610
|
APR, APR1, APS reductase 1, ATAPR1, PRH19, PAPS REDUCTASE HOMOLOG 19 |
-0.662 |
-0.993 |
| 449 |
264909_at |
AT2G17300
|
unknown |
-0.662 |
-0.422 |
| 450 |
253613_at |
AT4G30320
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.661 |
-0.262 |
| 451 |
267626_at |
AT2G42250
|
CYP712A1, cytochrome P450, family 712, subfamily A, polypeptide 1 |
-0.659 |
0.007 |
| 452 |
257204_at |
AT3G23805
|
RALFL24, ralf-like 24 |
-0.657 |
-0.318 |
| 453 |
256878_at |
AT3G26460
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.655 |
-0.743 |
| 454 |
256352_at |
AT1G54970
|
ATPRP1, proline-rich protein 1, PRP1, proline-rich protein 1, RHS7, ROOT HAIR SPECIFIC 7 |
-0.653 |
-0.096 |
| 455 |
249340_at |
AT5G41140
|
[Myosin heavy chain-related protein] |
-0.652 |
-0.062 |
| 456 |
252341_at |
AT3G48940
|
[Remorin family protein] |
-0.648 |
-0.049 |
| 457 |
263841_at |
AT2G36870
|
AtXTH32, XTH32, xyloglucan endotransglucosylase/hydrolase 32 |
-0.647 |
-0.537 |
| 458 |
253073_at |
AT4G37410
|
CYP81F4, cytochrome P450, family 81, subfamily F, polypeptide 4 |
-0.647 |
-0.272 |
| 459 |
251167_at |
AT3G63360
|
[defensin-related] |
-0.647 |
-0.077 |
| 460 |
253687_at |
AT4G29520
|
[LOCATED IN: endoplasmic reticulum, plasma membrane] |
-0.646 |
-0.272 |
| 461 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
-0.645 |
-0.463 |
| 462 |
267228_at |
AT2G43890
|
[Pectin lyase-like superfamily protein] |
-0.644 |
-0.073 |
| 463 |
265117_at |
AT1G62500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.641 |
0.071 |
| 464 |
257668_at |
AT3G20460
|
[Major facilitator superfamily protein] |
-0.641 |
-0.098 |
| 465 |
249202_at |
AT5G42580
|
CYP705A12, cytochrome P450, family 705, subfamily A, polypeptide 12 |
-0.640 |
-0.053 |
| 466 |
258912_at |
AT3G06460
|
[GNS1/SUR4 membrane protein family] |
-0.639 |
-0.348 |
| 467 |
264010_at |
AT2G21100
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.636 |
-0.339 |
| 468 |
252501_at |
AT3G46880
|
unknown |
-0.635 |
-0.313 |
| 469 |
264391_at |
AT1G11920
|
[Pectin lyase-like superfamily protein] |
-0.634 |
0.066 |
| 470 |
264495_at |
AT1G27380
|
RIC2, ROP-interactive CRIB motif-containing protein 2 |
-0.631 |
-0.110 |
| 471 |
263098_at |
AT2G16005
|
[MD-2-related lipid recognition domain-containing protein] |
-0.631 |
-0.164 |
| 472 |
258178_at |
AT3G21680
|
unknown |
-0.630 |
-0.220 |
| 473 |
248681_at |
AT5G48900
|
[Pectin lyase-like superfamily protein] |
-0.628 |
0.087 |
| 474 |
254634_at |
AT4G18650
|
[transcription factor-related] |
-0.626 |
0.693 |
| 475 |
248139_at |
AT5G54970
|
unknown |
-0.625 |
-0.417 |
| 476 |
252763_at |
AT3G42725
|
[Putative membrane lipoprotein] |
-0.624 |
-0.040 |
| 477 |
252238_at |
AT3G49960
|
[Peroxidase superfamily protein] |
-0.624 |
-0.211 |
| 478 |
254467_at |
AT4G20450
|
[Leucine-rich repeat protein kinase family protein] |
-0.623 |
-0.021 |
| 479 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-0.622 |
-0.291 |
| 480 |
263136_at |
AT1G78580
|
ATTPS1, trehalose-6-phosphate synthase, TPS1, trehalose-6-phosphate synthase, TPS1, TREHALOSE-6-PHOSPHATE SYNTHASE 1 |
-0.621 |
-0.294 |
| 481 |
258675_at |
AT3G08770
|
LTP6, lipid transfer protein 6 |
-0.620 |
-0.301 |
| 482 |
259763_at |
AT1G77630
|
LYM3, lysin-motif (LysM) domain protein 3, LYP3, LysM-containing receptor protein 3 |
-0.616 |
-0.353 |
| 483 |
250682_x_at |
AT5G06630
|
[proline-rich extensin-like family protein] |
-0.616 |
-0.278 |
| 484 |
248891_at |
AT5G46280
|
MCM3, MINICHROMOSOME MAINTENANCE 3 |
-0.616 |
-0.502 |
| 485 |
255011_at |
AT4G10040
|
CYTC-2, cytochrome c-2 |
-0.614 |
-0.083 |
| 486 |
259851_at |
AT1G72250
|
[Di-glucose binding protein with Kinesin motor domain] |
-0.614 |
-0.237 |
| 487 |
264565_at |
AT1G05280
|
unknown |
-0.613 |
-0.166 |
| 488 |
259850_at |
AT1G72240
|
unknown |
-0.613 |
0.015 |
| 489 |
253165_at |
AT4G35320
|
unknown |
-0.611 |
-0.481 |
| 490 |
256563_at |
AT3G29780
|
RALFL27, ralf-like 27 |
-0.610 |
-0.017 |
| 491 |
266649_at |
AT2G25810
|
TIP4;1, tonoplast intrinsic protein 4;1 |
-0.610 |
-0.317 |
| 492 |
253191_at |
AT4G35350
|
XCP1, xylem cysteine peptidase 1 |
-0.610 |
-0.112 |
| 493 |
247477_at |
AT5G62340
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.609 |
-0.116 |
| 494 |
266770_at |
AT2G03090
|
ATEXP15, ATEXPA15, expansin A15, ATHEXP ALPHA 1.3, EXP15, EXPANSIN 15, EXPA15, expansin A15 |
-0.609 |
-0.354 |
| 495 |
262659_at |
AT1G14240
|
[GDA1/CD39 nucleoside phosphatase family protein] |
-0.609 |
-0.116 |
| 496 |
249583_at |
AT5G37770
|
CML24, CALMODULIN-LIKE 24, TCH2, TOUCH 2 |
-0.608 |
-0.200 |
| 497 |
260547_at |
AT2G43550
|
[Scorpion toxin-like knottin superfamily protein] |
-0.607 |
-0.275 |
| 498 |
264114_at |
AT2G31270
|
ATCDT1A, ARABIDOPSIS HOMOLOG OF YEAST CDT1 A, CDT1, ARABIDOPSIS HOMOLOG OF YEAST CDT1, CDT1A, homolog of yeast CDT1 A |
-0.607 |
-0.368 |
| 499 |
256128_at |
AT1G18140
|
ATLAC1, LAC1, laccase 1 |
-0.606 |
-0.259 |
| 500 |
250434_at |
AT5G10390
|
H3.1, histone 3.1 |
-0.606 |
-0.462 |
| 501 |
254633_at |
AT4G18640
|
MRH1, morphogenesis of root hair 1 |
-0.603 |
-0.112 |
| 502 |
250173_at |
AT5G14340
|
AtMYB40, myb domain protein 40, MYB40, myb domain protein 40 |
-0.600 |
0.085 |
| 503 |
265586_at |
AT2G19990
|
PR-1-LIKE, pathogenesis-related protein-1-like |
-0.599 |
-0.054 |
| 504 |
261133_at |
AT1G19715
|
[Mannose-binding lectin superfamily protein] |
-0.597 |
0.124 |
| 505 |
261921_at |
AT1G65900
|
unknown |
-0.597 |
-0.200 |
| 506 |
258270_at |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
-0.595 |
-0.032 |
| 507 |
252302_at |
AT3G49190
|
[O-acyltransferase (WSD1-like) family protein] |
-0.594 |
-0.178 |
| 508 |
245546_at |
AT4G15290
|
ATCSLB05, ATCSLB5, CELLULOSE SYNTHASE LIKE 5, CSLB05 |
-0.593 |
-0.311 |
| 509 |
266684_at |
AT2G19720
|
rps15ab, ribosomal protein S15A B |
-0.593 |
-0.163 |
| 510 |
254313_at |
AT4G22460
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.591 |
-0.005 |
| 511 |
255460_at |
AT4G02800
|
unknown |
-0.590 |
-0.421 |
| 512 |
260135_at |
AT1G66400
|
CML23, calmodulin like 23 |
-0.590 |
0.170 |
| 513 |
250933_at |
AT5G03170
|
ATFLA11, ARABIDOPSIS FASCICLIN-LIKE ARABINOGALACTAN-PROTEIN 11, FLA11, FASCICLIN-like arabinogalactan-protein 11 |
-0.587 |
-0.311 |
| 514 |
258832_at |
AT3G07070
|
[Protein kinase superfamily protein] |
-0.586 |
-0.143 |
| 515 |
267457_at |
AT2G33790
|
AGP30, arabinogalactan protein 30, ATAGP30 |
-0.586 |
-0.033 |
| 516 |
255934_at |
AT1G12740
|
CYP87A2, cytochrome P450, family 87, subfamily A, polypeptide 2 |
-0.584 |
-0.245 |
| 517 |
264573_at |
AT1G05310
|
[Pectin lyase-like superfamily protein] |
-0.583 |
-0.204 |
| 518 |
255005_at |
AT4G09990
|
GXM2, glucuronoxylan methyltransferase 2 |
-0.583 |
-0.184 |
| 519 |
262608_at |
AT1G14120
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.580 |
-0.319 |
| 520 |
246401_at |
AT1G57560
|
AtMYB50, myb domain protein 50, MYB50, myb domain protein 50 |
-0.580 |
-0.121 |
| 521 |
264262_at |
AT1G09200
|
H3.1, histone 3.1 |
-0.578 |
-0.299 |
| 522 |
259655_at |
AT1G55210
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.577 |
-0.651 |
| 523 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
-0.577 |
-0.809 |
| 524 |
257896_at |
AT3G16920
|
ATCTL2, CTL2, chitinase-like protein 2 |
-0.576 |
-0.010 |
| 525 |
257481_at |
AT1G08430
|
ALMT1, aluminum-activated malate transporter 1, ATALMT1, ARABIDOPSIS THALIANA ALUMINUM-ACTIVATED MALATE TRANSPORTER 1 |
-0.576 |
-0.211 |
| 526 |
252594_at |
AT3G45680
|
[Major facilitator superfamily protein] |
-0.576 |
-0.234 |
| 527 |
254332_at |
AT4G22730
|
[Leucine-rich repeat protein kinase family protein] |
-0.575 |
-0.293 |
| 528 |
252692_at |
AT3G43960
|
[Cysteine proteinases superfamily protein] |
-0.575 |
-0.172 |
| 529 |
249718_at |
AT5G35740
|
[Carbohydrate-binding X8 domain superfamily protein] |
-0.574 |
-0.116 |
| 530 |
249832_at |
AT5G23400
|
[Leucine-rich repeat (LRR) family protein] |
-0.573 |
-0.097 |
| 531 |
261777_at |
AT1G76210
|
unknown |
-0.572 |
-0.012 |
| 532 |
251857_at |
AT3G54770
|
ARP1, ABA-regulated RNA-binding protein 1 |
-0.572 |
-0.117 |
| 533 |
251281_at |
AT3G61640
|
AGP20, arabinogalactan protein 20, AtAGP20, Arabinogalactan protein 20 |
-0.571 |
-0.207 |
| 534 |
258529_at |
AT3G06740
|
GATA15, GATA transcription factor 15 |
-0.570 |
-0.118 |
| 535 |
266271_at |
AT2G29440
|
ATGSTU6, glutathione S-transferase tau 6, GST24, GLUTATHIONE S-TRANSFERASE 24, GSTU6, glutathione S-transferase tau 6 |
-0.568 |
-0.430 |
| 536 |
249920_at |
AT5G19260
|
FAF3, FANTASTIC FOUR 3 |
-0.566 |
-0.391 |
| 537 |
249392_at |
AT5G40150
|
[Peroxidase superfamily protein] |
-0.566 |
-0.194 |
| 538 |
261653_at |
AT1G01900
|
ATSBT1.1, SBTI1.1 |
-0.566 |
-0.452 |
| 539 |
261203_at |
AT1G12845
|
unknown |
-0.565 |
0.002 |
| 540 |
266326_at |
AT2G46650
|
ATCB5-C, ARABIDOPSIS CYTOCHROME B5 ISOFORM C, B5 #1, CB5-C, cytochrome B5 isoform C, CYTB5-C |
-0.564 |
-0.769 |
| 541 |
251843_x_at |
AT3G54590
|
ATHRGP1, hydroxyproline-rich glycoprotein, HRGP1, hydroxyproline-rich glycoprotein |
-0.564 |
-0.286 |
| 542 |
249881_at |
AT5G23190
|
CYP86B1, cytochrome P450, family 86, subfamily B, polypeptide 1 |
-0.562 |
-0.199 |
| 543 |
261330_at |
AT1G44900
|
ATMCM2, MCM2, MINICHROMOSOME MAINTENANCE 2 |
-0.562 |
-0.426 |
| 544 |
257151_at |
AT3G27200
|
[Cupredoxin superfamily protein] |
-0.561 |
-0.201 |
| 545 |
261460_at |
AT1G07880
|
ATMPK13 |
-0.560 |
-0.179 |
| 546 |
264287_at |
AT1G61930
|
unknown |
-0.560 |
-0.163 |
| 547 |
247638_at |
AT5G60490
|
AtFLA12, FLA12, FASCICLIN-like arabinogalactan-protein 12 |
-0.559 |
-0.377 |
| 548 |
262148_at |
AT1G52560
|
[HSP20-like chaperones superfamily protein] |
-0.558 |
-0.176 |
| 549 |
263628_at |
AT2G04780
|
FLA7, FASCICLIN-like arabinoogalactan 7 |
-0.553 |
-0.307 |
| 550 |
261550_at |
AT1G63450
|
RHS8, root hair specific 8, XUT1, XYLOGLUCAN-SPECIFIC GALACTURONOSYLTRANSFERASE 1 |
-0.553 |
-0.288 |
| 551 |
247581_at |
AT5G61350
|
[Protein kinase superfamily protein] |
-0.553 |
-0.191 |
| 552 |
265204_at |
AT2G36650
|
unknown |
-0.552 |
-0.044 |
| 553 |
267372_at |
AT2G26290
|
ARSK1, root-specific kinase 1 |
-0.548 |
-0.147 |
| 554 |
267094_at |
AT2G38080
|
ATLMCO4, ARABIDOPSIS LACCASE-LIKE MULTICOPPER OXIDASE 4, IRX12, IRREGULAR XYLEM 12, LAC4, LACCASE 4, LMCO4, LACCASE-LIKE MULTICOPPER OXIDASE 4 |
-0.546 |
-0.051 |
| 555 |
260366_at |
AT1G70460
|
AtPERK13, proline-rich extensin-like receptor kinase 13, PERK13, proline-rich extensin-like receptor kinase 13, RHS10, root hair specific 10 |
-0.543 |
-0.222 |
| 556 |
266489_at |
AT2G35190
|
ATNPSN11, NPSN11, novel plant snare 11, NSPN11 |
-0.543 |
-0.112 |
| 557 |
257824_at |
AT3G25290
|
[Auxin-responsive family protein] |
-0.542 |
-0.238 |
| 558 |
254076_at |
AT4G25340
|
ATFKBP53, FKBP53, FK506 BINDING PROTEIN 53 |
-0.541 |
-0.377 |
| 559 |
245133_at |
AT2G45310
|
GAE4, UDP-D-glucuronate 4-epimerase 4 |
-0.540 |
-0.218 |
| 560 |
260150_at |
AT1G52820
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.540 |
0.021 |
| 561 |
253353_at |
AT4G33730
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.539 |
-0.205 |
| 562 |
247030_at |
AT5G67210
|
IRX15-L, IRX15-LIKE |
-0.538 |
-0.146 |
| 563 |
245980_at |
AT5G13140
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.538 |
-0.189 |
| 564 |
258145_at |
AT3G18200
|
UMAMIT4, Usually multiple acids move in and out Transporters 4 |
-0.537 |
-0.158 |
| 565 |
250778_at |
AT5G05500
|
MOP10 |
-0.537 |
-0.527 |
| 566 |
258474_at |
AT3G02650
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.537 |
-0.325 |
| 567 |
251908_at |
AT3G53770
|
[late embryogenesis abundant 3 (LEA3) family protein] |
-0.535 |
0.001 |
| 568 |
248252_at |
AT5G53250
|
AGP22, arabinogalactan protein 22, ATAGP22, ARABINOGALACTAN PROTEIN 22 |
-0.535 |
-0.136 |
| 569 |
245912_at |
AT5G19600
|
SULTR3;5, sulfate transporter 3;5 |
-0.535 |
-0.199 |
| 570 |
261807_at |
AT1G30515
|
unknown |
-0.535 |
0.037 |
| 571 |
267137_at |
AT2G23410
|
ACPT, cis-prenyltransferase, AtcPT3, CPT, cis-prenyltransferase, cPT3, cis-prenyltransferase 3 |
-0.533 |
-0.062 |
| 572 |
266264_at |
AT2G27775
|
unknown |
-0.533 |
-0.233 |
| 573 |
247406_at |
AT5G62920
|
ARR6, response regulator 6 |
-0.532 |
-0.066 |
| 574 |
267169_at |
AT2G37540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.531 |
-0.138 |
| 575 |
262109_at |
AT1G02730
|
ATCSLD5, cellulose synthase-like D5, CSLD5, CELLULOSE SYNTHASE LIKE D5, CSLD5, cellulose synthase-like D5, SOS6, SALT OVERLY SENSITIVE 6 |
-0.531 |
-0.483 |
| 576 |
250471_at |
AT5G10170
|
ATMIPS3, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 3, MIPS3, myo-inositol-1-phosphate synthase 3 |
-0.529 |
-0.033 |
| 577 |
264315_at |
AT1G70370
|
PG2, polygalacturonase 2 |
-0.529 |
-0.317 |
| 578 |
252711_at |
AT3G43720
|
LTPG2, glycosylphosphatidylinositol-anchored lipid protein transfer 2 |
-0.527 |
-0.314 |
| 579 |
259801_at |
AT1G72230
|
[Cupredoxin superfamily protein] |
-0.527 |
-0.231 |
| 580 |
250958_at |
AT5G03260
|
LAC11, laccase 11 |
-0.526 |
-0.240 |
| 581 |
260645_at |
AT1G53300
|
TTL1, tetratricopeptide-repeat thioredoxin-like 1 |
-0.526 |
-0.164 |
| 582 |
247727_at |
AT5G59490
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.526 |
-0.151 |
| 583 |
261080_at |
AT1G07370
|
ATPCNA1, PROLIFERATING CELLULAR NUCLEAR ANTIGEN 1, PCNA1, proliferating cellular nuclear antigen 1 |
-0.525 |
-0.289 |
| 584 |
254044_at |
AT4G25820
|
ATXTH14, XTH14, xyloglucan endotransglucosylase/hydrolase 14, XTR9, xyloglucan endotransglycosylase 9 |
-0.525 |
-0.527 |
| 585 |
257724_at |
AT3G18510
|
unknown |
-0.524 |
-0.185 |
| 586 |
252442_at |
AT3G46940
|
DUT1, DUTP-PYROPHOSPHATASE-LIKE 1 |
-0.524 |
-0.399 |
| 587 |
249848_at |
AT5G23220
|
NIC3, nicotinamidase 3 |
-0.524 |
-0.256 |
| 588 |
258222_at |
AT3G15680
|
[Ran BP2/NZF zinc finger-like superfamily protein] |
-0.522 |
-0.543 |
| 589 |
261898_at |
AT1G80720
|
[Mitochondrial glycoprotein family protein] |
-0.522 |
-0.287 |
| 590 |
252078_at |
AT3G51740
|
IMK2, inflorescence meristem receptor-like kinase 2 |
-0.522 |
-0.200 |
| 591 |
254181_at |
AT4G23940
|
ARC1, ACCUMULATION AND REPLICATION OF CHLOROPLASTS 1, FtsHi1, FTSH inactive protease 1 |
-0.521 |
-0.238 |
| 592 |
258480_at |
AT3G02640
|
unknown |
-0.521 |
-0.385 |
| 593 |
252833_at |
AT4G40090
|
AGP3, arabinogalactan protein 3 |
-0.520 |
-0.337 |
| 594 |
248150_at |
AT5G54670
|
ATK3, kinesin 3, KATC, KINESIN-LIKE PROTEIN IN ARABIDOPSIS THALIANA C |
-0.520 |
-0.530 |
| 595 |
245584_at |
AT4G14940
|
AO1, amine oxidase 1, ATAO1, amine oxidase 1 |
-0.520 |
-0.350 |
| 596 |
264184_at |
AT1G54790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.519 |
-0.195 |
| 597 |
259346_at |
AT3G03910
|
glutamate dehydrogenase 3 |
-0.518 |
-0.106 |
| 598 |
254125_at |
AT4G24670
|
TAR2, tryptophan aminotransferase related 2 |
-0.517 |
-0.135 |
| 599 |
251584_at |
AT3G58620
|
TTL4, tetratricopetide-repeat thioredoxin-like 4 |
-0.516 |
0.150 |
| 600 |
249151_at |
AT5G43360
|
ATPT4, PHT1;3, phosphate transporter 1;3, PHT3, PHOSPHATE TRANSPORTER 3 |
-0.515 |
-0.045 |