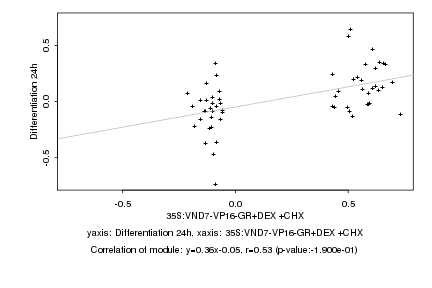
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
35S:VND7-VP16-GR+DEX +CHX |
Log2 signal ratio
Differentiation 24h |
| 1 |
256018_at |
AT1G58300
|
HO4, heme oxygenase 4 |
0.730 |
-0.110 |
| 2 |
249439_at |
AT5G40020
|
[Pathogenesis-related thaumatin superfamily protein] |
0.696 |
0.178 |
| 3 |
247590_at |
AT5G60720
|
unknown |
0.663 |
0.331 |
| 4 |
250322_at |
AT5G12870
|
ATMYB46, MYB DOMAIN PROTEIN 46, MYB46, myb domain protein 46 |
0.652 |
0.346 |
| 5 |
266713_at |
AT2G46760
|
AtGulLO6, GulLO6, L -gulono-1,4-lactone ( L -GulL) oxidase 6 |
0.650 |
0.128 |
| 6 |
246827_at |
AT5G26330
|
[Cupredoxin superfamily protein] |
0.636 |
0.350 |
| 7 |
245584_at |
AT4G14940
|
AO1, amine oxidase 1, ATAO1, amine oxidase 1 |
0.633 |
0.100 |
| 8 |
249227_at |
AT5G42180
|
PER64, peroxidase 64 |
0.620 |
0.137 |
| 9 |
250622_at |
AT5G07310
|
[Integrase-type DNA-binding superfamily protein] |
0.617 |
0.300 |
| 10 |
265672_at |
AT2G31980
|
AtCYS2, PHYTOCYSTATIN 2, CYS2, PHYTOCYSTATIN 2 |
0.604 |
0.123 |
| 11 |
251992_at |
AT3G53350
|
MIDD1, Microtubule Depletion Domain 1, RIP3, ROP interactive partner 3, RIP4, ROP interactive partner 4 |
0.604 |
0.466 |
| 12 |
247269_at |
AT5G64210
|
AOX2, alternative oxidase 2 |
0.592 |
-0.012 |
| 13 |
252006_at |
AT3G52820
|
ATPAP22, PURPLE ACID PHOSPHATASE 22, PAP22, purple acid phosphatase 22 |
0.588 |
-0.025 |
| 14 |
254674_at |
AT4G18450
|
[Integrase-type DNA-binding superfamily protein] |
0.586 |
0.077 |
| 15 |
245697_at |
AT5G04200
|
AtMC9, metacaspase 9, AtMCP2f, metacaspase 2f, MC9, metacaspase 9, MCP2f, metacaspase 2f |
0.585 |
-0.025 |
| 16 |
252439_at |
AT3G47400
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.576 |
0.337 |
| 17 |
253786_at |
AT4G28650
|
[Leucine-rich repeat transmembrane protein kinase family protein] |
0.561 |
0.113 |
| 18 |
249518_at |
AT5G38610
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.558 |
0.193 |
| 19 |
265024_at |
AT1G24600
|
unknown |
0.540 |
0.218 |
| 20 |
261546_at |
AT1G63520
|
unknown |
0.519 |
0.198 |
| 21 |
261845_at |
AT1G15960
|
ATNRAMP6, NRAMP6, NRAMP metal ion transporter 6 |
0.516 |
-0.133 |
| 22 |
252025_at |
AT3G52900
|
unknown |
0.510 |
0.643 |
| 23 |
251069_at |
AT5G01930
|
AtMAN6, MAN6, endo-beta-mannase 6 |
0.503 |
-0.086 |
| 24 |
260333_at |
AT1G70500
|
[Pectin lyase-like superfamily protein] |
0.501 |
0.579 |
| 25 |
248998_at |
AT5G45320
|
unknown |
0.494 |
-0.047 |
| 26 |
247965_at |
AT5G56540
|
AGP14, arabinogalactan protein 14, ATAGP14 |
0.454 |
0.093 |
| 27 |
259268_at |
AT3G01070
|
AtENODL16, ENODL16, early nodulin-like protein 16 |
0.440 |
0.050 |
| 28 |
246138_at |
AT5G19870
|
unknown |
0.435 |
-0.048 |
| 29 |
255701_at |
AT4G00220
|
JLO, JAGGED LATERAL ORGANS, LBD30, LOB DOMAIN-CONTAINING PROTEIN 30 |
0.429 |
-0.042 |
| 30 |
251093_at |
AT5G01360
|
TBL3, TRICHOME BIREFRINGENCE-LIKE 3 |
0.427 |
0.241 |
| 31 |
251181_at |
AT3G62820
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.213 |
0.078 |
| 32 |
256499_at |
AT1G36640
|
unknown |
-0.192 |
-0.039 |
| 33 |
251509_at |
AT3G59010
|
PME35, pectin methylesterase 35, PME61, pectin methylesterase 61 |
-0.183 |
-0.221 |
| 34 |
248317_at |
AT5G52680
|
[Copper transport protein family] |
-0.158 |
0.015 |
| 35 |
262682_at |
AT1G75900
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.155 |
-0.157 |
| 36 |
259429_at |
AT1G01600
|
CYP86A4, cytochrome P450, family 86, subfamily A, polypeptide 4 |
-0.138 |
-0.083 |
| 37 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
-0.135 |
-0.370 |
| 38 |
255266_at |
AT4G05200
|
CRK25, cysteine-rich RLK (RECEPTOR-like protein kinase) 25 |
-0.133 |
-0.088 |
| 39 |
252510_at |
AT3G46270
|
[receptor protein kinase-related] |
-0.131 |
0.016 |
| 40 |
254663_at |
AT4G18290
|
KAT2, potassium channel in Arabidopsis thaliana 2 |
-0.129 |
0.163 |
| 41 |
259371_at |
AT1G69080
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.118 |
-0.234 |
| 42 |
252340_at |
AT3G48920
|
AtMYB45, myb domain protein 45, MYB45, myb domain protein 45 |
-0.114 |
-0.059 |
| 43 |
261429_at |
AT1G18860
|
ATWRKY61, WRKY DNA-BINDING PROTEIN 61, WRKY61, WRKY DNA-binding protein 61 |
-0.109 |
-0.135 |
| 44 |
254202_at |
AT4G24140
|
[alpha/beta-Hydrolases superfamily protein] |
-0.107 |
-0.224 |
| 45 |
265186_at |
AT1G23560
|
unknown |
-0.106 |
-0.015 |
| 46 |
265842_at |
AT2G35700
|
ATERF38, ERF FAMILY PROTEIN 38, ERF38, ERF family protein 38 |
-0.105 |
0.040 |
| 47 |
256359_at |
AT1G66460
|
[Protein kinase superfamily protein] |
-0.104 |
-0.086 |
| 48 |
263830_at |
AT2G40260
|
[Homeodomain-like superfamily protein] |
-0.100 |
-0.470 |
| 49 |
257673_at |
AT3G20370
|
[TRAF-like family protein] |
-0.092 |
0.345 |
| 50 |
255732_at |
AT1G25450
|
CER60, ECERIFERUM 60, KCS5, 3-ketoacyl-CoA synthase 5 |
-0.090 |
-0.730 |
| 51 |
255766_at |
AT1G16750
|
unknown |
-0.086 |
-0.358 |
| 52 |
266669_at |
AT2G29750
|
UGT71C1, UDP-glucosyl transferase 71C1 |
-0.086 |
-0.042 |
| 53 |
254340_at |
AT4G22120
|
[ERD (early-responsive to dehydration stress) family protein] |
-0.084 |
0.237 |
| 54 |
259527_at |
AT1G12600
|
[UDP-N-acetylglucosamine (UAA) transporter family] |
-0.074 |
0.019 |
| 55 |
251477_at |
AT3G59680
|
unknown |
-0.071 |
0.090 |
| 56 |
266917_at |
AT2G45830
|
DTA2, downstream target of AGL15 2 |
-0.070 |
-0.152 |
| 57 |
251000_at |
AT5G02650
|
unknown |
-0.067 |
-0.010 |
| 58 |
263081_at |
AT2G27220
|
BLH5, BEL1-like homeodomain 5 |
-0.061 |
-0.092 |
| 59 |
250299_at |
AT5G11910
|
[alpha/beta-Hydrolases superfamily protein] |
-0.059 |
-0.079 |