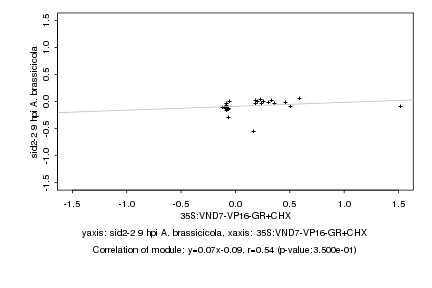
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
35S:VND7-VP16-GR+CHX |
Log2 signal ratio
sid2-2 9 hpi A. brassicicola |
| 1 |
260173_at |
AT1G71930
|
ANAC030, Arabidopsis NAC domain containing protein 30, VND7, vascular related NAC-domain protein 7 |
1.515 |
-0.081 |
| 2 |
260333_at |
AT1G70500
|
[Pectin lyase-like superfamily protein] |
0.583 |
0.062 |
| 3 |
263629_at |
AT2G04850
|
[Auxin-responsive family protein] |
0.505 |
-0.091 |
| 4 |
253191_at |
AT4G35350
|
XCP1, xylem cysteine peptidase 1 |
0.459 |
-0.010 |
| 5 |
251297_at |
AT3G62020
|
GLP10, germin-like protein 10 |
0.353 |
-0.031 |
| 6 |
251093_at |
AT5G01360
|
TBL3, TRICHOME BIREFRINGENCE-LIKE 3 |
0.331 |
0.023 |
| 7 |
252006_at |
AT3G52820
|
ATPAP22, PURPLE ACID PHOSPHATASE 22, PAP22, purple acid phosphatase 22 |
0.296 |
-0.009 |
| 8 |
259963_at |
AT1G53660
|
[nodulin MtN21 /EamA-like transporter family protein] |
0.250 |
0.007 |
| 9 |
265672_at |
AT2G31980
|
AtCYS2, PHYTOCYSTATIN 2, CYS2, PHYTOCYSTATIN 2 |
0.234 |
-0.033 |
| 10 |
251739_at |
AT3G56170
|
CAN, Ca-2+ dependent nuclease, CAN1, calcium dependent nuclease 1 |
0.224 |
0.050 |
| 11 |
247269_at |
AT5G64210
|
AOX2, alternative oxidase 2 |
0.195 |
0.002 |
| 12 |
256803_at |
AT3G20960
|
CYP705A33, cytochrome P450, family 705, subfamily A, polypeptide 33 |
0.184 |
-0.027 |
| 13 |
261399_at |
AT1G79620
|
[Leucine-rich repeat protein kinase family protein] |
0.180 |
0.020 |
| 14 |
265161_at |
AT1G30900
|
BP80-3;3, binding protein of 80 kDa 3;3, VSR3;3, VACUOLAR SORTING RECEPTOR 3;3, VSR6, VACUOLAR SORTING RECEPTOR 6 |
0.162 |
-0.538 |
| 15 |
257772_at |
AT3G23080
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.124 |
-0.102 |
| 16 |
245947_at |
AT5G19530
|
ACL5, ACAULIS 5 |
-0.099 |
-0.102 |
| 17 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.099 |
-0.075 |
| 18 |
258256_at |
AT3G26890
|
unknown |
-0.095 |
-0.108 |
| 19 |
250017_at |
AT5G18140
|
DJC69, DNA J protein A69 |
-0.095 |
-0.122 |
| 20 |
248541_at |
AT5G50180
|
[Protein kinase superfamily protein] |
-0.087 |
-0.151 |
| 21 |
247278_at |
AT5G64380
|
[Inositol monophosphatase family protein] |
-0.087 |
-0.022 |
| 22 |
265060_at |
AT1G52150
|
ATHB-15, ATHB15, CNA, CORONA, ICU4, INCURVATA 4 |
-0.084 |
-0.065 |
| 23 |
249709_at |
AT5G35670
|
iqd33, IQ-domain 33 |
-0.080 |
-0.126 |
| 24 |
267535_at |
AT2G41940
|
ZFP8, zinc finger protein 8 |
-0.071 |
-0.284 |
| 25 |
265967_at |
AT2G37450
|
UMAMIT13, Usually multiple acids move in and out Transporters 13 |
-0.068 |
-0.111 |
| 26 |
252975_s_at |
AT4G38430
|
ATROPGEF1, ROPGEF1, ROP (rho of plants) guanine nucleotide exchange factor 1 |
-0.066 |
-0.136 |
| 27 |
264007_at |
AT2G21140
|
ATPRP2, proline-rich protein 2, PRP2, proline-rich protein 2 |
-0.063 |
0.003 |