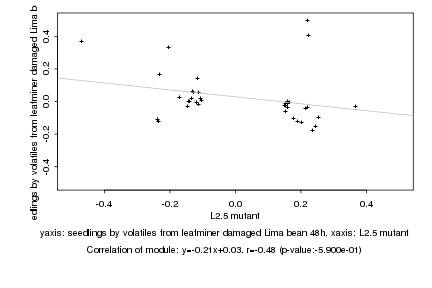
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
L2.5 mutant |
Log2 signal ratio
seedlings by volatiles from leafminer damaged Lima bean 48h |
| 1 |
248178_at |
AT5G54370
|
[Late embryogenesis abundant (LEA) protein-related] |
0.365 |
-0.029 |
| 2 |
265050_at |
AT1G52070
|
[Mannose-binding lectin superfamily protein] |
0.251 |
-0.092 |
| 3 |
247605_at |
AT5G60970
|
TCP5, TEOSINTE BRANCHED 1, cycloidea and PCF transcription factor 5 |
0.244 |
-0.152 |
| 4 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
0.234 |
-0.175 |
| 5 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
0.221 |
0.408 |
| 6 |
266154_at |
AT2G12190
|
[Cytochrome P450 superfamily protein] |
0.219 |
-0.032 |
| 7 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.218 |
0.503 |
| 8 |
267271_at |
AT2G02540
|
ATHB21, homeobox protein 21, HB21, homeobox protein 21, ZFHD4, ZINC FINGER HOMEODOMAIN 4, ZHD3, ZINC FINGER HOMEODOMAIN 3 |
0.214 |
-0.039 |
| 9 |
252073_at |
AT3G51750
|
unknown |
0.201 |
-0.125 |
| 10 |
263115_at |
AT1G03055
|
AtD27, A. thaliana homolog of rice D27, D27, DWARF27 |
0.187 |
-0.118 |
| 11 |
250161_at |
AT5G15240
|
[Transmembrane amino acid transporter family protein] |
0.176 |
-0.102 |
| 12 |
249518_at |
AT5G38610
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.165 |
-0.000 |
| 13 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
0.159 |
-0.011 |
| 14 |
255023_at |
AT4G09850
|
unknown |
0.158 |
0.003 |
| 15 |
254173_at |
AT4G24580
|
REN1, ROP1 ENHANCER 1 |
0.157 |
-0.032 |
| 16 |
254482_at |
AT4G20370
|
TSF, TWIN SISTER OF FT |
0.152 |
-0.007 |
| 17 |
248394_at |
AT5G52070
|
[Agenet domain-containing protein] |
0.151 |
-0.058 |
| 18 |
245245_at |
AT1G44318
|
hemb2 |
0.150 |
-0.022 |
| 19 |
250574_at |
AT5G08230
|
[Tudor/PWWP/MBT domain-containing protein] |
0.148 |
-0.020 |
| 20 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
-0.471 |
0.372 |
| 21 |
245049_at |
ATCG00050
|
RPS16, ribosomal protein S16 |
-0.241 |
-0.106 |
| 22 |
263605_at |
AT2G16485
|
NERD, Needed for RDR2-independent DNA methylation |
-0.238 |
-0.120 |
| 23 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.235 |
0.170 |
| 24 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
-0.206 |
0.333 |
| 25 |
250245_at |
AT5G13690
|
CYL1, CYCLOPS 1, NAGLU, N-ACETYL-GLUCOSAMINIDASE |
-0.172 |
0.026 |
| 26 |
248599_at |
AT5G49380
|
unknown |
-0.149 |
-0.029 |
| 27 |
262829_at |
AT1G14970
|
[O-fucosyltransferase family protein] |
-0.145 |
0.004 |
| 28 |
264948_at |
AT1G77030
|
[hydrolases, acting on acid anhydrides, in phosphorus-containing anhydrides] |
-0.141 |
0.002 |
| 29 |
248704_at |
AT5G48450
|
sks3, SKU5 similar 3 |
-0.135 |
0.022 |
| 30 |
245156_at |
AT5G12480
|
CPK7, calmodulin-domain protein kinase 7 |
-0.134 |
0.063 |
| 31 |
266360_at |
AT2G32250
|
FRS2, FAR1-related sequence 2 |
-0.129 |
0.056 |
| 32 |
252431_at |
AT3G47700
|
MAG2, maigo2 |
-0.119 |
0.000 |
| 33 |
246765_at |
AT5G27330
|
[Prefoldin chaperone subunit family protein] |
-0.117 |
0.147 |
| 34 |
253432_at |
AT4G32450
|
MEF8S, MEF8 similar |
-0.115 |
-0.017 |
| 35 |
248744_at |
AT5G48250
|
BBX8, B-box domain protein 8 |
-0.113 |
0.058 |
| 36 |
260658_at |
AT1G19410
|
[FBD / Leucine Rich Repeat domains containing protein] |
-0.108 |
0.022 |
| 37 |
258781_at |
AT3G11740
|
unknown |
-0.105 |
0.011 |