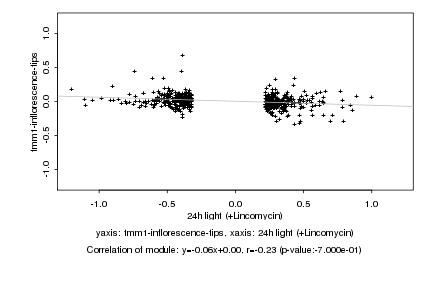
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
24h light (+Lincomycin) |
Log2 signal ratio
tmm1-inflorescence-tips |
| 1 |
256459_at |
AT1G36180
|
ACC2, acetyl-CoA carboxylase 2 |
0.993 |
0.066 |
| 2 |
263515_at |
AT2G21640
|
unknown |
0.884 |
0.082 |
| 3 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
0.854 |
-0.126 |
| 4 |
261222_at |
AT1G20120
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.837 |
-0.048 |
| 5 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.788 |
-0.293 |
| 6 |
251871_at |
AT3G54520
|
unknown |
0.784 |
0.021 |
| 7 |
258327_at |
AT3G22640
|
PAP85 |
0.780 |
-0.076 |
| 8 |
255893_at |
AT1G17960
|
[Threonyl-tRNA synthetase] |
0.768 |
0.162 |
| 9 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.708 |
-0.192 |
| 10 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
0.697 |
-0.289 |
| 11 |
247615_at |
AT5G60250
|
[zinc finger (C3HC4-type RING finger) family protein] |
0.657 |
0.162 |
| 12 |
260668_at |
AT1G19530
|
unknown |
0.645 |
-0.028 |
| 13 |
247317_at |
AT5G64060
|
anac103, NAC domain containing protein 103, NAC103, NAC domain containing protein 103 |
0.644 |
-0.005 |
| 14 |
259982_at |
AT1G76410
|
ATL8 |
0.644 |
-0.201 |
| 15 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
0.641 |
0.067 |
| 16 |
264124_at |
AT1G79360
|
ATOCT2, organic cation/carnitine transporter 2, OCT2, ORGANIC CATION TRANSPORTER 2, OCT2, organic cation/carnitine transporter 2 |
0.635 |
0.007 |
| 17 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.621 |
0.135 |
| 18 |
256188_at |
AT1G30160
|
unknown |
0.613 |
0.015 |
| 19 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
0.610 |
-0.055 |
| 20 |
245233_at |
AT4G25580
|
[CAP160 protein] |
0.589 |
0.124 |
| 21 |
249784_at |
AT5G24280
|
GMI1, GAMMA-IRRADIATION AND MITOMYCIN C INDUCED 1 |
0.582 |
-0.049 |
| 22 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
0.563 |
-0.060 |
| 23 |
247951_at |
AT5G57240
|
ORP4C, OSBP(oxysterol binding protein)-related protein 4C |
0.561 |
0.076 |
| 24 |
251282_at |
AT3G61630
|
CRF6, cytokinin response factor 6 |
0.559 |
-0.068 |
| 25 |
252019_at |
AT3G53040
|
[late embryogenesis abundant protein, putative / LEA protein, putative] |
0.558 |
0.052 |
| 26 |
264774_at |
AT1G22890
|
unknown |
0.558 |
-0.202 |
| 27 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
0.557 |
-0.123 |
| 28 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.554 |
-0.011 |
| 29 |
255941_at |
AT1G20350
|
ATTIM17-1, translocase inner membrane subunit 17-1, TIM17-1, translocase inner membrane subunit 17-1 |
0.538 |
0.024 |
| 30 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
0.525 |
0.016 |
| 31 |
262347_at |
AT1G64110
|
DAA1, DUO1-activated ATPase 1 |
0.517 |
0.095 |
| 32 |
251455_at |
AT3G60100
|
CSY5, citrate synthase 5 |
0.516 |
-0.028 |
| 33 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
0.506 |
0.152 |
| 34 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
0.499 |
-0.081 |
| 35 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.497 |
0.013 |
| 36 |
261192_at |
AT1G32870
|
ANAC013, Arabidopsis NAC domain containing protein 13, ANAC13, NAC domain protein 13, NAC13, NAC domain protein 13 |
0.493 |
0.036 |
| 37 |
245392_at |
AT4G15680
|
[Thioredoxin superfamily protein] |
0.483 |
-0.059 |
| 38 |
256114_at |
AT1G16850
|
unknown |
0.479 |
0.085 |
| 39 |
247339_at |
AT5G63690
|
[Nucleic acid-binding, OB-fold-like protein] |
0.474 |
-0.089 |
| 40 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
0.473 |
-0.026 |
| 41 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
0.473 |
-0.286 |
| 42 |
252539_at |
AT3G45730
|
unknown |
0.472 |
0.019 |
| 43 |
264238_at |
AT1G54740
|
unknown |
0.469 |
-0.312 |
| 44 |
267246_at |
AT2G30250
|
ATWRKY25, WRKY25, WRKY DNA-binding protein 25 |
0.469 |
0.032 |
| 45 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.466 |
-0.018 |
| 46 |
255223_at |
AT4G05370
|
[BCS1 AAA-type ATPase] |
0.466 |
0.033 |
| 47 |
254193_at |
AT4G23870
|
unknown |
0.463 |
-0.194 |
| 48 |
259409_at |
AT1G13330
|
AHP2, Arabidopsis Hop2 homolog |
0.461 |
-0.009 |
| 49 |
250182_at |
AT5G14470
|
AtGALK2, GALK2, galactokinase 2 |
0.433 |
-0.059 |
| 50 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.433 |
0.348 |
| 51 |
248260_at |
AT5G53240
|
unknown |
0.432 |
-0.004 |
| 52 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
0.427 |
-0.327 |
| 53 |
257667_at |
AT3G20440
|
BE1, BRANCHING ENZYME 1, EMB2729, EMBRYO DEFECTIVE 2729 |
0.426 |
-0.025 |
| 54 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
0.424 |
0.044 |
| 55 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
0.420 |
0.250 |
| 56 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
0.418 |
-0.000 |
| 57 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
0.411 |
0.039 |
| 58 |
256970_at |
AT3G21090
|
ABCG15, ATP-binding cassette G15 |
0.409 |
-0.031 |
| 59 |
248611_at |
AT5G49520
|
ATWRKY48, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 48, WRKY48, WRKY DNA-binding protein 48 |
0.406 |
-0.072 |
| 60 |
265926_at |
AT2G18600
|
[Ubiquitin-conjugating enzyme family protein] |
0.405 |
-0.009 |
| 61 |
266503_at |
AT2G47780
|
[Rubber elongation factor protein (REF)] |
0.404 |
0.078 |
| 62 |
253051_at |
AT4G37490
|
CYC1, CYCLIN 1, CYCB1, CYCB1;1, CYCLIN B1;1 |
0.400 |
0.049 |
| 63 |
250365_at |
AT5G11410
|
[Protein kinase superfamily protein] |
0.395 |
0.029 |
| 64 |
253975_at |
AT4G26600
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.389 |
0.014 |
| 65 |
250504_at |
AT5G09840
|
[Putative endonuclease or glycosyl hydrolase] |
0.388 |
-0.038 |
| 66 |
264433_at |
AT1G61810
|
BGLU45, beta-glucosidase 45 |
0.388 |
-0.200 |
| 67 |
250420_at |
AT5G11260
|
HY5, ELONGATED HYPOCOTYL 5, TED 5, REVERSAL OF THE DET PHENOTYPE 5 |
0.387 |
-0.039 |
| 68 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
0.387 |
0.045 |
| 69 |
245970_at |
AT5G20710
|
BGAL7, beta-galactosidase 7 |
0.381 |
0.051 |
| 70 |
266511_at |
AT2G47680
|
[zinc finger (CCCH type) helicase family protein] |
0.381 |
0.017 |
| 71 |
250666_at |
AT5G07100
|
WRKY26, WRKY DNA-binding protein 26 |
0.380 |
0.060 |
| 72 |
252330_at |
AT3G48770
|
[DNA binding] |
0.379 |
-0.010 |
| 73 |
261243_at |
AT1G20180
|
unknown |
0.378 |
0.012 |
| 74 |
260522_x_at |
AT2G41730
|
unknown |
0.377 |
0.138 |
| 75 |
264167_at |
AT1G02060
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.377 |
-0.218 |
| 76 |
254573_at |
AT4G19420
|
[Pectinacetylesterase family protein] |
0.374 |
0.011 |
| 77 |
246700_at |
AT5G28030
|
DES1, L-cysteine desulfhydrase 1 |
0.373 |
0.106 |
| 78 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
0.373 |
-0.138 |
| 79 |
267011_at |
AT2G39230
|
LOJ, LATERAL ORGAN JUNCTION |
0.369 |
-0.069 |
| 80 |
258347_at |
AT3G17520
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.367 |
0.088 |
| 81 |
255500_at |
AT4G02390
|
APP, poly(ADP-ribose) polymerase, PARP2, poly(ADP-ribose) polymerase 2, PP, poly(ADP-ribose) polymerase |
0.365 |
0.066 |
| 82 |
250914_at |
AT5G03780
|
TRFL10, TRF-like 10 |
0.364 |
-0.059 |
| 83 |
253019_at |
AT4G38010
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.364 |
-0.130 |
| 84 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.361 |
0.067 |
| 85 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
0.361 |
0.073 |
| 86 |
249619_at |
AT5G37500
|
GORK, gated outwardly-rectifying K+ channel |
0.359 |
-0.089 |
| 87 |
246251_at |
AT4G37220
|
[Cold acclimation protein WCOR413 family] |
0.359 |
-0.041 |
| 88 |
254015_at |
AT4G26140
|
BGAL12, beta-galactosidase 12 |
0.359 |
-0.026 |
| 89 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.355 |
0.089 |
| 90 |
249575_at |
AT5G37670
|
[HSP20-like chaperones superfamily protein] |
0.354 |
-0.039 |
| 91 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.352 |
-0.070 |
| 92 |
250680_at |
AT5G06570
|
[alpha/beta-Hydrolases superfamily protein] |
0.351 |
0.016 |
| 93 |
255742_at |
AT1G25560
|
AtTEM1, EDF1, ETHYLENE RESPONSE DNA BINDING FACTOR 1, TEM1, TEMPRANILLO 1 |
0.351 |
-0.112 |
| 94 |
248895_at |
AT5G46330
|
FLS2, FLAGELLIN-SENSITIVE 2 |
0.349 |
-0.136 |
| 95 |
264375_at |
AT2G25090
|
AtCIPK16, CIPK16, CBL-interacting protein kinase 16, SnRK3.18, SNF1-RELATED PROTEIN KINASE 3.18 |
0.348 |
0.049 |
| 96 |
262586_at |
AT1G15480
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.346 |
-0.086 |
| 97 |
251975_at |
AT3G53230
|
AtCDC48B, cell division cycle 48B |
0.345 |
0.018 |
| 98 |
248347_at |
AT5G52250
|
EFO1, EARLY FLOWERING BY OVEREXPRESSION 1, RUP1, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 |
0.343 |
-0.028 |
| 99 |
260641_at |
AT1G53200
|
unknown |
0.343 |
-0.116 |
| 100 |
248821_at |
AT5G47070
|
[Protein kinase superfamily protein] |
0.341 |
0.033 |
| 101 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.341 |
-0.022 |
| 102 |
246461_at |
AT5G16930
|
[AAA-type ATPase family protein] |
0.335 |
-0.030 |
| 103 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
0.334 |
-0.095 |
| 104 |
253829_at |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
0.333 |
-0.172 |
| 105 |
252281_at |
AT3G49320
|
[Metal-dependent protein hydrolase] |
0.331 |
-0.087 |
| 106 |
266518_at |
AT2G35170
|
[Histone H3 K4-specific methyltransferase SET7/9 family protein] |
0.330 |
0.008 |
| 107 |
262801_at |
AT1G21010
|
unknown |
0.325 |
-0.040 |
| 108 |
247515_at |
AT5G61740
|
ABCA10, ATP-binding cassette A10, ATATH14, ARABIDOPSIS THALIANA ABC2 HOMOLOG 14, ATH14, ABC2 homolog 14 |
0.323 |
-0.012 |
| 109 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
0.323 |
-0.255 |
| 110 |
266479_at |
AT2G31160
|
LSH3, LIGHT SENSITIVE HYPOCOTYLS 3, OBO1, ORGAN BOUNDARY 1 |
0.323 |
-0.026 |
| 111 |
250196_at |
AT5G14580
|
[polyribonucleotide nucleotidyltransferase, putative] |
0.322 |
-0.013 |
| 112 |
266849_at |
AT2G25940
|
ALPHA-VPE, alpha-vacuolar processing enzyme, ALPHAVPE |
0.321 |
-0.029 |
| 113 |
257395_at |
AT2G15630
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.318 |
-0.056 |
| 114 |
263118_at |
AT1G03090
|
MCCA |
0.317 |
-0.137 |
| 115 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.316 |
0.021 |
| 116 |
264522_at |
AT1G10050
|
[glycosyl hydrolase family 10 protein / carbohydrate-binding domain-containing protein] |
0.313 |
-0.057 |
| 117 |
255317_at |
AT4G04180
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.310 |
0.033 |
| 118 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
0.310 |
0.007 |
| 119 |
246850_at |
AT5G26860
|
LON1, lon protease 1, LON_ARA_ARA |
0.308 |
0.049 |
| 120 |
258235_at |
AT3G27620
|
AOX1C, alternative oxidase 1C |
0.307 |
-0.053 |
| 121 |
260741_at |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
0.307 |
0.120 |
| 122 |
253872_at |
AT4G27410
|
ANAC072, Arabidopsis NAC domain containing protein 72, RD26, RESPONSIVE TO DESICCATION 26 |
0.305 |
0.049 |
| 123 |
258545_at |
AT3G07050
|
NSN1, nucleostemin-like 1 |
0.305 |
-0.056 |
| 124 |
249019_at |
AT5G44780
|
MORF4, multiple organellar RNA editing factor 4 |
0.304 |
0.142 |
| 125 |
259793_at |
AT1G64380
|
[Integrase-type DNA-binding superfamily protein] |
0.303 |
-0.031 |
| 126 |
267337_at |
AT2G39980
|
[HXXXD-type acyl-transferase family protein] |
0.303 |
0.020 |
| 127 |
262395_at |
AT1G49540
|
AtELP2, Elongator subunit 2, ELP2, elongator protein 2 |
0.302 |
-0.000 |
| 128 |
265061_at |
AT1G61640
|
[Protein kinase superfamily protein] |
0.301 |
0.056 |
| 129 |
247855_at |
AT5G58210
|
[hydroxyproline-rich glycoprotein family protein] |
0.301 |
0.004 |
| 130 |
247954_at |
AT5G56870
|
BGAL4, beta-galactosidase 4 |
0.301 |
-0.288 |
| 131 |
253388_at |
AT4G32910
|
unknown |
0.299 |
-0.028 |
| 132 |
251554_at |
AT3G58670
|
unknown |
0.297 |
-0.087 |
| 133 |
257522_at |
AT3G08990
|
[Yippee family putative zinc-binding protein] |
0.297 |
-0.097 |
| 134 |
258227_at |
AT3G15620
|
UVR3, UV REPAIR DEFECTIVE 3 |
0.297 |
-0.100 |
| 135 |
264175_at |
AT1G02050
|
LAP6, LESS ADHESIVE POLLEN 6, PSKA, polyketide synthase A |
0.295 |
-0.069 |
| 136 |
247015_at |
AT5G66960
|
[Prolyl oligopeptidase family protein] |
0.294 |
-0.081 |
| 137 |
264379_at |
AT2G25200
|
unknown |
0.294 |
0.009 |
| 138 |
248668_at |
AT5G48720
|
XRI, X-RAY INDUCED TRANSCRIPT, XRI1, X-RAY INDUCED TRANSCRIPT 1 |
0.294 |
-0.008 |
| 139 |
262456_at |
AT1G11260
|
ATSTP1, SUGAR TRANSPORTER 1, STP1, sugar transporter 1 |
0.293 |
-0.043 |
| 140 |
254079_at |
AT4G25730
|
[FtsJ-like methyltransferase family protein] |
0.292 |
-0.046 |
| 141 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.292 |
0.072 |
| 142 |
262552_at |
AT1G31350
|
KUF1, KAR-UP F-box 1 |
0.292 |
0.028 |
| 143 |
264027_at |
AT2G03670
|
CDC48B, cell division cycle 48B |
0.291 |
-0.083 |
| 144 |
262255_at |
AT1G53790
|
[F-box and associated interaction domains-containing protein] |
0.290 |
-0.029 |
| 145 |
267414_at |
AT2G34790
|
EDA28, EMBRYO SAC DEVELOPMENT ARREST 28, MEE23, MATERNAL EFFECT EMBRYO ARREST 23 |
0.289 |
0.181 |
| 146 |
256107_at |
AT1G16830
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.288 |
-0.206 |
| 147 |
261065_at |
AT1G07500
|
SMR5, SIAMESE-RELATED 5 |
0.288 |
0.129 |
| 148 |
245528_at |
AT4G15530
|
PPDK, pyruvate orthophosphate dikinase |
0.288 |
0.326 |
| 149 |
259579_at |
AT1G28010
|
ABCB14, ATP-binding cassette B14, ATABCB14, Arabidopsis thaliana ATP-binding cassette B14, MDR12, multi-drug resistance 12, PGP14, P-glycoprotein 14 |
0.287 |
-0.027 |
| 150 |
251373_at |
AT3G60530
|
GATA4, GATA transcription factor 4 |
0.286 |
-0.099 |
| 151 |
249426_at |
AT5G39840
|
[ATP-dependent RNA helicase, mitochondrial, putative] |
0.285 |
-0.062 |
| 152 |
254197_at |
AT4G24040
|
ATTRE1, TRE1, trehalase 1 |
0.285 |
-0.038 |
| 153 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
0.285 |
-0.094 |
| 154 |
248984_at |
AT5G45140
|
NRPC2, nuclear RNA polymerase C2 |
0.284 |
0.014 |
| 155 |
256340_at |
AT1G72070
|
[Chaperone DnaJ-domain superfamily protein] |
0.283 |
-0.012 |
| 156 |
264752_at |
AT1G23010
|
LPR1, Low Phosphate Root1 |
0.282 |
0.089 |
| 157 |
252834_at |
AT4G40070
|
[RING/U-box superfamily protein] |
0.282 |
-0.019 |
| 158 |
248869_at |
AT5G46840
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.282 |
-0.083 |
| 159 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.278 |
0.106 |
| 160 |
267035_at |
AT2G38400
|
AGT3, alanine:glyoxylate aminotransferase 3 |
0.278 |
-0.097 |
| 161 |
262624_at |
AT1G06450
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
0.278 |
-0.070 |
| 162 |
259606_at |
AT1G27920
|
MAP65-8, microtubule-associated protein 65-8 |
0.278 |
-0.005 |
| 163 |
259076_at |
AT3G02140
|
AFP4, ABI FIVE BINDING PROTEIN 4, TMAC2, TWO OR MORE ABRES-CONTAINING GENE 2 |
0.278 |
0.004 |
| 164 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
0.278 |
-0.032 |
| 165 |
255278_at |
AT4G04940
|
[transducin family protein / WD-40 repeat family protein] |
0.277 |
0.065 |
| 166 |
256024_at |
AT1G58340
|
BCD1, BUSH-AND-CHLOROTIC-DWARF 1, ZF14, ZRZ, ZRIZI |
0.277 |
0.026 |
| 167 |
254933_at |
AT4G11130
|
RDR2, RNA-dependent RNA polymerase 2, SMD1, SILENCING MOVEMENT DEFICIENT 1 |
0.275 |
0.004 |
| 168 |
262548_at |
AT1G31280
|
AGO2, argonaute 2, AtAGO2 |
0.274 |
-0.058 |
| 169 |
246582_at |
AT1G31750
|
[proline-rich family protein] |
0.274 |
0.066 |
| 170 |
265274_at |
AT2G28450
|
[zinc finger (CCCH-type) family protein] |
0.274 |
-0.053 |
| 171 |
266532_at |
AT2G16890
|
[UDP-Glycosyltransferase superfamily protein] |
0.273 |
0.113 |
| 172 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
0.271 |
0.188 |
| 173 |
265588_at |
AT2G19970
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.270 |
-0.194 |
| 174 |
257035_at |
AT3G19270
|
CYP707A4, cytochrome P450, family 707, subfamily A, polypeptide 4 |
0.270 |
0.137 |
| 175 |
246595_at |
AT5G14780
|
FDH, formate dehydrogenase |
0.270 |
0.061 |
| 176 |
250024_at |
AT5G18270
|
ANAC087, Arabidopsis NAC domain containing protein 87 |
0.269 |
-0.042 |
| 177 |
260272_at |
AT1G80570
|
[RNI-like superfamily protein] |
0.269 |
-0.011 |
| 178 |
251356_at |
AT3G61060
|
AtPP2-A13, phloem protein 2-A13, PP2-A13, phloem protein 2-A13 |
0.268 |
-0.049 |
| 179 |
255521_at |
AT4G02280
|
ATSUS3, SUS3, sucrose synthase 3 |
0.267 |
0.107 |
| 180 |
261654_at |
AT1G01920
|
[SET domain-containing protein] |
0.267 |
-0.072 |
| 181 |
262336_at |
AT1G64220
|
TOM7-2, translocase of outer membrane 7 kDa subunit 2 |
0.266 |
-0.151 |
| 182 |
258117_at |
AT3G14700
|
[SART-1 family] |
0.266 |
0.015 |
| 183 |
256413_at |
AT3G11100
|
[sequence-specific DNA binding transcription factors] |
0.266 |
-0.045 |
| 184 |
266732_at |
AT2G03240
|
[EXS (ERD1/XPR1/SYG1) family protein] |
0.265 |
-0.035 |
| 185 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
0.262 |
-0.081 |
| 186 |
260296_at |
AT1G63750
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.262 |
0.045 |
| 187 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
0.262 |
-0.094 |
| 188 |
251650_at |
AT3G57360
|
unknown |
0.261 |
-0.165 |
| 189 |
248307_at |
AT5G52850
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.261 |
-0.009 |
| 190 |
248723_at |
AT5G47950
|
[HXXXD-type acyl-transferase family protein] |
0.261 |
0.014 |
| 191 |
257288_at |
AT3G29670
|
PMAT2, phenolic glucoside malonyltransferase 2 |
0.260 |
0.037 |
| 192 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
0.260 |
0.109 |
| 193 |
255872_at |
AT2G30360
|
CIPK11, CBL-INTERACTING PROTEIN KINASE 11, PKS5, PROTEIN KINASE SOS2-LIKE 5, SIP4, SOS3-interacting protein 4, SNRK3.22, SNF1-RELATED PROTEIN KINASE 3.22 |
0.259 |
-0.081 |
| 194 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
0.259 |
0.078 |
| 195 |
256097_at |
AT1G13670
|
unknown |
0.259 |
-0.165 |
| 196 |
265116_at |
AT1G62480
|
[Vacuolar calcium-binding protein-related] |
0.259 |
-0.004 |
| 197 |
264482_at |
AT1G77210
|
AtSTP14, sugar transport protein 14, STP14, sugar transport protein 14 |
0.257 |
-0.002 |
| 198 |
250506_at |
AT5G09930
|
ABCF2, ATP-binding cassette F2 |
0.257 |
0.012 |
| 199 |
263707_at |
AT1G09300
|
[Metallopeptidase M24 family protein] |
0.256 |
-0.007 |
| 200 |
246600_at |
AT5G14930
|
SAG101, senescence-associated gene 101 |
0.256 |
-0.001 |
| 201 |
255479_at |
AT4G02380
|
AtLEA5, Arabidopsis thaliana late embryogenensis abundant like 5, SAG21, senescence-associated gene 21 |
0.256 |
0.066 |
| 202 |
254055_at |
AT4G25330
|
unknown |
0.255 |
0.049 |
| 203 |
245523_at |
AT4G15910
|
ATDI21, drought-induced 21, DI21, drought-induced 21 |
0.252 |
-0.040 |
| 204 |
261937_at |
AT1G22570
|
[Major facilitator superfamily protein] |
0.252 |
-0.164 |
| 205 |
252488_at |
AT3G46700
|
[UDP-Glycosyltransferase superfamily protein] |
0.251 |
0.024 |
| 206 |
248060_at |
AT5G55560
|
[Protein kinase superfamily protein] |
0.250 |
-0.050 |
| 207 |
253322_at |
AT4G33980
|
unknown |
0.250 |
-0.157 |
| 208 |
263881_at |
AT2G21820
|
unknown |
0.250 |
0.049 |
| 209 |
247130_at |
AT5G66180
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.249 |
-0.019 |
| 210 |
245518_at |
AT4G15850
|
ATRH1, RNA helicase 1, RH1, RNA helicase 1 |
0.248 |
-0.045 |
| 211 |
263498_at |
AT2G42610
|
LSH10, LIGHT SENSITIVE HYPOCOTYLS 10 |
0.247 |
0.088 |
| 212 |
247676_at |
AT5G59980
|
AtRPP30, GAF1, GAMETOPHYTE DEFECTIVE 1, RPP30 |
0.247 |
-0.032 |
| 213 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
0.247 |
0.065 |
| 214 |
248231_at |
AT5G53770
|
[Nucleotidyltransferase family protein] |
0.246 |
0.018 |
| 215 |
252508_at |
AT3G46210
|
[Ribosomal protein S5 domain 2-like superfamily protein] |
0.245 |
-0.036 |
| 216 |
255430_at |
AT4G03320
|
AtTic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV, Tic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV |
0.245 |
0.059 |
| 217 |
246238_at |
AT4G36670
|
AtPLT6, AtPMT6, PLT6, polyol transporter 6, PMT6, polyol/monosaccharide transporter 6 |
0.245 |
0.246 |
| 218 |
264265_at |
AT1G09280
|
unknown |
0.245 |
-0.059 |
| 219 |
253492_at |
AT4G31810
|
[ATP-dependent caseinolytic (Clp) protease/crotonase family protein] |
0.244 |
0.041 |
| 220 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.244 |
0.040 |
| 221 |
254018_at |
AT4G26180
|
CoAc2, CoA Carrier 2 |
0.244 |
0.032 |
| 222 |
262618_at |
AT1G06560
|
[NOL1/NOP2/sun family protein] |
0.244 |
-0.010 |
| 223 |
266695_at |
AT2G19810
|
AtOZF1, AtTZF2, OZF1, Oxidation-related Zinc Finger 1, TZF2, tandem zinc finger 2 |
0.244 |
-0.036 |
| 224 |
258546_at |
AT3G07060
|
emb1974, embryo defective 1974 |
0.243 |
-0.055 |
| 225 |
259285_at |
AT3G11460
|
MEF10, mitochondrial RNA editing factor 10 |
0.243 |
0.060 |
| 226 |
249839_at |
AT5G23405
|
[HMG-box (high mobility group) DNA-binding family protein] |
0.243 |
-0.114 |
| 227 |
261524_at |
AT1G14300
|
[ARM repeat superfamily protein] |
0.243 |
-0.029 |
| 228 |
263847_at |
AT2G36970
|
[UDP-Glycosyltransferase superfamily protein] |
0.242 |
0.014 |
| 229 |
259915_at |
AT1G72790
|
[hydroxyproline-rich glycoprotein family protein] |
0.241 |
0.036 |
| 230 |
258374_at |
AT3G14360
|
[alpha/beta-Hydrolases superfamily protein] |
0.241 |
-0.037 |
| 231 |
246550_at |
AT5G14920
|
GASA14, A-stimulated in Arabidopsis 14 |
0.241 |
-0.054 |
| 232 |
261586_at |
AT1G01640
|
[BTB/POZ domain-containing protein] |
0.241 |
0.036 |
| 233 |
261310_at |
AT1G05750
|
CLB19, PDE247, pigment defective 247 |
0.240 |
-0.041 |
| 234 |
257679_at |
AT3G20470
|
ATGRP-5, ATGRP5, GRP-5, GLYCINE-RICH PROTEIN 5, GRP5, glycine-rich protein 5 |
0.240 |
-0.001 |
| 235 |
259831_at |
AT1G69600
|
ATHB29, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 29, ZFHD1, zinc finger homeodomain 1, ZHD11, ZINC FINGER HOMEODOMAIN 11 |
0.239 |
0.020 |
| 236 |
261086_at |
AT1G17460
|
TRFL3, TRF-like 3 |
0.239 |
-0.113 |
| 237 |
249128_at |
AT5G43440
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.239 |
-0.076 |
| 238 |
266471_at |
AT2G31060
|
EMB2785, EMBRYO DEFECTIVE 2785 |
0.238 |
-0.013 |
| 239 |
252386_at |
AT3G47840
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.238 |
-0.098 |
| 240 |
254778_at |
AT4G12750
|
[Homeodomain-like transcriptional regulator] |
0.237 |
-0.099 |
| 241 |
259444_at |
AT1G02370
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.236 |
-0.076 |
| 242 |
265931_at |
AT2G18520
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.236 |
0.104 |
| 243 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
0.235 |
-0.003 |
| 244 |
263067_at |
AT2G17550
|
TRM26, TON1 Recruiting Motif 26 |
0.235 |
-0.136 |
| 245 |
246034_at |
AT5G08350
|
[GRAM domain-containing protein / ABA-responsive protein-related] |
0.235 |
-0.075 |
| 246 |
266140_at |
AT2G28120
|
[Major facilitator superfamily protein] |
0.235 |
-0.044 |
| 247 |
256697_at |
AT3G20660
|
AtOCT4, organic cation/carnitine transporter4, OCT4, organic cation/carnitine transporter4 |
0.235 |
-0.011 |
| 248 |
253240_at |
AT4G34510
|
KCS17, 3-ketoacyl-CoA synthase 17 |
0.235 |
0.051 |
| 249 |
260774_at |
AT1G78290
|
SNRK2-8, SNF1-RELATED PROTEIN KINASE 2-8, SNRK2.8, SNF1-RELATED PROTEIN KINASE 2.8, SRK2C, SNF1-RELATED PROTEIN KINASE 2C |
0.234 |
0.077 |
| 250 |
258476_at |
AT3G02400
|
[SMAD/FHA domain-containing protein ] |
0.234 |
-0.026 |
| 251 |
252770_at |
AT3G42860
|
[zinc knuckle (CCHC-type) family protein] |
0.234 |
0.023 |
| 252 |
263320_at |
AT2G47180
|
AtGolS1, galactinol synthase 1, GolS1, galactinol synthase 1 |
0.233 |
0.003 |
| 253 |
263196_at |
AT1G36070
|
[Transducin/WD40 repeat-like superfamily protein] |
0.233 |
-0.096 |
| 254 |
258026_at |
AT3G19290
|
ABF4, ABRE binding factor 4, AREB2, ABA-RESPONSIVE ELEMENT BINDING PROTEIN 2 |
0.232 |
-0.050 |
| 255 |
251432_at |
AT3G59820
|
AtLETM1, LETM1, leucine zipper-EF-hand-containing transmembrane protein 1 |
0.232 |
-0.065 |
| 256 |
247129_at |
AT5G66150
|
[Glycosyl hydrolase family 38 protein] |
0.231 |
0.034 |
| 257 |
246906_at |
AT5G25475
|
[AP2/B3-like transcriptional factor family protein] |
0.231 |
-0.029 |
| 258 |
265373_at |
AT2G06510
|
ATRPA1A, ARABIDOPSIS THALIANA REPLICATION PROTEIN A 1A, ATRPA70A, ARABIDOPSIS THALIANA RPA70-KDA SUBUNIT A, RPA1A, replication protein A 1A, RPA70A, RPA70-KDA SUBUNIT A |
0.231 |
-0.092 |
| 259 |
264513_at |
AT1G09420
|
G6PD4, glucose-6-phosphate dehydrogenase 4 |
0.231 |
0.015 |
| 260 |
247277_at |
AT5G64420
|
[DNA polymerase V family] |
0.230 |
-0.017 |
| 261 |
250029_at |
AT5G18200
|
[UTP:galactose-1-phosphate uridylyltransferases] |
0.230 |
-0.045 |
| 262 |
267568_at |
AT2G30780
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.229 |
-0.092 |
| 263 |
251229_at |
AT3G62740
|
BGLU7, beta glucosidase 7 |
0.228 |
0.005 |
| 264 |
249624_at |
AT5G37570
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.228 |
0.025 |
| 265 |
261070_at |
AT1G07390
|
AtRLP1, receptor like protein 1, RLP1, receptor like protein 1 |
0.228 |
-0.019 |
| 266 |
245137_at |
AT2G45460
|
[SMAD/FHA domain-containing protein ] |
0.227 |
-0.003 |
| 267 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
0.227 |
-0.076 |
| 268 |
257460_at |
AT1G75580
|
SAUR51, SMALL AUXIN UPREGULATED RNA 51 |
0.227 |
-0.069 |
| 269 |
264251_at |
AT1G09190
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.226 |
-0.010 |
| 270 |
262092_at |
AT1G56150
|
SAUR71, SMALL AUXIN UPREGULATED 71 |
0.226 |
-0.012 |
| 271 |
252192_at |
AT3G50000
|
ATCKA2, CKA2, casein kinase II, alpha chain 2 |
0.225 |
-0.034 |
| 272 |
250032_at |
AT5G18170
|
GDH1, glutamate dehydrogenase 1 |
0.225 |
-0.031 |
| 273 |
258953_at |
AT3G01430
|
unknown |
0.223 |
0.200 |
| 274 |
253258_at |
AT4G34400
|
[AP2/B3-like transcriptional factor family protein] |
0.223 |
-0.053 |
| 275 |
259058_at |
AT3G03470
|
CYP89A9, cytochrome P450, family 87, subfamily A, polypeptide 9 |
0.222 |
0.047 |
| 276 |
259364_at |
AT1G13260
|
EDF4, ETHYLENE RESPONSE DNA BINDING FACTOR 4, RAV1, related to ABI3/VP1 1 |
0.222 |
-0.115 |
| 277 |
246951_at |
AT5G04880
|
[expressed protein] |
0.222 |
-0.070 |
| 278 |
252355_at |
AT3G48250
|
BIR6, Buthionine sulfoximine-insensitive roots 6 |
0.222 |
-0.062 |
| 279 |
258718_at |
AT3G09760
|
[RING/U-box superfamily protein] |
0.221 |
-0.036 |
| 280 |
245543_at |
AT4G15260
|
[UDP-Glycosyltransferase superfamily protein] |
0.221 |
0.039 |
| 281 |
264637_at |
AT1G65560
|
[Zinc-binding dehydrogenase family protein] |
0.221 |
-0.085 |
| 282 |
254174_at |
AT4G24120
|
ATYSL1, YELLOW STRIPE LIKE 1, YSL1, YELLOW STRIPE like 1 |
0.221 |
-0.030 |
| 283 |
264561_at |
AT1G55810
|
UKL3, uridine kinase-like 3 |
0.220 |
-0.040 |
| 284 |
245558_at |
AT4G15430
|
[ERD (early-responsive to dehydration stress) family protein] |
0.220 |
0.144 |
| 285 |
257968_at |
AT3G27550
|
[RNA-binding CRS1 / YhbY (CRM) domain protein] |
0.220 |
0.003 |
| 286 |
247449_at |
AT5G62290
|
[nucleotide-sensitive chloride conductance regulator (ICln) family protein] |
0.220 |
-0.031 |
| 287 |
267411_at |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
0.219 |
-0.068 |
| 288 |
249806_at |
AT5G23850
|
unknown |
0.219 |
0.051 |
| 289 |
254854_at |
AT4G12130
|
[Glycine cleavage T-protein family] |
0.218 |
0.090 |
| 290 |
262659_at |
AT1G14240
|
[GDA1/CD39 nucleoside phosphatase family protein] |
0.218 |
-0.043 |
| 291 |
248870_at |
AT5G46710
|
[PLATZ transcription factor family protein] |
0.218 |
-0.038 |
| 292 |
253777_at |
AT4G28450
|
[nucleotide binding] |
0.217 |
-0.077 |
| 293 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-1.208 |
0.189 |
| 294 |
255016_at |
AT4G10120
|
ATSPS4F |
-1.109 |
0.032 |
| 295 |
256096_at |
AT1G13650
|
unknown |
-1.101 |
-0.055 |
| 296 |
256603_at |
AT3G28270
|
unknown |
-1.054 |
0.018 |
| 297 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.983 |
0.058 |
| 298 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-0.919 |
0.028 |
| 299 |
254250_at |
AT4G23290
|
CRK21, cysteine-rich RLK (RECEPTOR-like protein kinase) 21 |
-0.908 |
0.226 |
| 300 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
-0.898 |
0.025 |
| 301 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
-0.860 |
0.042 |
| 302 |
260877_at |
AT1G21500
|
unknown |
-0.836 |
-0.017 |
| 303 |
249732_at |
AT5G24420
|
PGL5, 6-phosphogluconolactonase 5 |
-0.811 |
0.003 |
| 304 |
258302_at |
AT3G30610
|
unknown |
-0.806 |
-0.018 |
| 305 |
260380_at |
AT1G73870
|
BBX16, B-box domain protein 16, COL7, CONSTANS-LIKE 7 |
-0.779 |
0.118 |
| 306 |
258055_at |
AT3G16250
|
NDF4, NDH-dependent cyclic electron flow 1, PnsB3, Photosynthetic NDH subcomplex B 3 |
-0.778 |
-0.006 |
| 307 |
259161_at |
AT3G01500
|
ATBCA1, BETA CARBONIC ANHYDRASE 1, ATSABP3, ARABIDOPSIS THALIANA SALICYLIC ACID-BINDING PROTEIN 3, CA1, carbonic anhydrase 1, SABP3, SALICYLIC ACID-BINDING PROTEIN 3 |
-0.751 |
-0.041 |
| 308 |
252618_at |
AT3G45140
|
ATLOX2, ARABIODOPSIS THALIANA LIPOXYGENASE 2, LOX2, lipoxygenase 2 |
-0.741 |
0.450 |
| 309 |
251142_at |
AT5G01015
|
unknown |
-0.735 |
0.012 |
| 310 |
256527_at |
AT1G66100
|
[Plant thionin] |
-0.734 |
0.084 |
| 311 |
262309_at |
AT1G70820
|
[phosphoglucomutase, putative / glucose phosphomutase, putative] |
-0.709 |
-0.079 |
| 312 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
-0.706 |
0.009 |
| 313 |
253871_at |
AT4G27440
|
PORB, protochlorophyllide oxidoreductase B |
-0.691 |
-0.047 |
| 314 |
261769_at |
AT1G76100
|
PETE1, plastocyanin 1 |
-0.677 |
0.000 |
| 315 |
261942_at |
AT1G22590
|
AGL87, AGAMOUS-like 87 |
-0.675 |
0.131 |
| 316 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
-0.675 |
0.130 |
| 317 |
264037_at |
AT2G03750
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.664 |
0.010 |
| 318 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
-0.663 |
-0.064 |
| 319 |
252853_at |
AT4G39710
|
FKBP16-2, FK506-binding protein 16-2, PnsL4, Photosynthetic NDH subcomplex L 4 |
-0.654 |
-0.026 |
| 320 |
263114_at |
AT1G03130
|
PSAD-2, photosystem I subunit D-2 |
-0.640 |
0.004 |
| 321 |
246792_at |
AT5G27290
|
unknown |
-0.623 |
-0.033 |
| 322 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
-0.613 |
0.346 |
| 323 |
255248_at |
AT4G05180
|
PSBQ, PHOTOSYSTEM II SUBUNIT Q, PSBQ-2, photosystem II subunit Q-2, PSII-Q |
-0.610 |
0.024 |
| 324 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
-0.608 |
-0.080 |
| 325 |
256599_at |
AT3G14760
|
unknown |
-0.605 |
0.142 |
| 326 |
265611_at |
AT2G25510
|
unknown |
-0.601 |
0.005 |
| 327 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
-0.595 |
-0.033 |
| 328 |
261431_at |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
-0.594 |
-0.046 |
| 329 |
258025_at |
AT3G19480
|
3-PGDH, PGDH3, phosphoglycerate dehydrogenase 3 |
-0.593 |
0.042 |
| 330 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
-0.588 |
0.031 |
| 331 |
247100_at |
AT5G66520
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.582 |
-0.026 |
| 332 |
255088_at |
AT4G09350
|
CRRJ, CHLORORESPIRATORY REDUCTION J, DJC75, DNA J protein C75, NdhT, NADH dehydrogenase-like complex T |
-0.581 |
-0.029 |
| 333 |
265722_at |
AT2G40100
|
LHCB4.3, light harvesting complex photosystem II |
-0.573 |
0.018 |
| 334 |
258409_at |
AT3G17640
|
[Leucine-rich repeat (LRR) family protein] |
-0.572 |
0.048 |
| 335 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
-0.572 |
0.062 |
| 336 |
263715_at |
AT2G20570
|
GBF's pro-rich region-interacting factor 1 |
-0.567 |
0.049 |
| 337 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-0.566 |
0.149 |
| 338 |
249727_at |
AT5G35490
|
ATMRU1, ARABIDOPSIS MTO 1 RESPONDING UP 1, MRU1, MTO 1 RESPONDING UP 1 |
-0.564 |
0.053 |
| 339 |
256304_at |
AT1G69523
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.559 |
0.123 |
| 340 |
260547_at |
AT2G43550
|
[Scorpion toxin-like knottin superfamily protein] |
-0.558 |
0.015 |
| 341 |
263597_at |
AT2G01870
|
unknown |
-0.544 |
-0.072 |
| 342 |
246004_at |
AT5G20630
|
ATGER3, ARABIDOPSIS THALIANA GERMIN 3, GER3, germin 3, GLP3, GERMIN-LIKE PROTEIN 3, GLP3A, GLP3B |
-0.543 |
0.044 |
| 343 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
-0.543 |
0.091 |
| 344 |
262373_at |
AT1G73120
|
unknown |
-0.540 |
0.106 |
| 345 |
261474_at |
AT1G14540
|
PER4, peroxidase 4 |
-0.535 |
0.054 |
| 346 |
257793_at |
AT3G26960
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.534 |
0.348 |
| 347 |
247415_at |
AT5G63060
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.530 |
-0.002 |
| 348 |
261638_at |
AT1G49975
|
[INVOLVED IN: photosynthesis] |
-0.526 |
-0.018 |
| 349 |
262177_at |
AT1G74710
|
ATICS1, ARABIDOPSIS ISOCHORISMATE SYNTHASE 1, EDS16, ENHANCED DISEASE SUSCEPTIBILITY TO ERYSIPHE ORONTII 16, ICS1, ISOCHORISMATE SYNTHASE 1, SID2, SALICYLIC ACID INDUCTION DEFICIENT 2 |
-0.526 |
0.101 |
| 350 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
-0.525 |
0.093 |
| 351 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
-0.525 |
0.056 |
| 352 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.523 |
-0.012 |
| 353 |
261913_at |
AT1G65860
|
FMO GS-OX1, flavin-monooxygenase glucosinolate S-oxygenase 1 |
-0.523 |
0.203 |
| 354 |
247055_at |
AT5G66740
|
unknown |
-0.522 |
0.115 |
| 355 |
254233_at |
AT4G23800
|
3xHMG-box2, 3xHigh Mobility Group-box2 |
-0.521 |
-0.013 |
| 356 |
260431_at |
AT1G68190
|
BBX27, B-box domain protein 27 |
-0.520 |
-0.048 |
| 357 |
256655_at |
AT3G18890
|
AtTic62, translocon at the inner envelope membrane of chloroplasts 62, Tic62, translocon at the inner envelope membrane of chloroplasts 62 |
-0.513 |
0.016 |
| 358 |
256427_at |
AT3G11090
|
LBD21, LOB domain-containing protein 21 |
-0.509 |
0.007 |
| 359 |
250560_at |
AT5G08020
|
ATRPA70B, ARABIDOPSIS THALIANA RPA70-KDA SUBUNIT B, RPA70B, RPA70-kDa subunit B |
-0.506 |
0.080 |
| 360 |
261046_at |
AT1G01390
|
[UDP-Glycosyltransferase superfamily protein] |
-0.506 |
0.011 |
| 361 |
259188_at |
AT3G01510
|
LSF1, like SEX4 1 |
-0.505 |
0.045 |
| 362 |
258322_at |
AT3G22740
|
HMT3, homocysteine S-methyltransferase 3 |
-0.505 |
0.125 |
| 363 |
248807_at |
AT5G47500
|
PME5, pectin methylesterase 5 |
-0.504 |
0.012 |
| 364 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
-0.502 |
0.024 |
| 365 |
255046_at |
AT4G09650
|
ATPD, ATP synthase delta-subunit gene, PDE332, PIGMENT DEFECTIVE 332 |
-0.499 |
-0.014 |
| 366 |
264839_at |
AT1G03630
|
POR C, protochlorophyllide oxidoreductase C, PORC |
-0.498 |
0.043 |
| 367 |
245343_at |
AT4G15830
|
[ARM repeat superfamily protein] |
-0.494 |
0.072 |
| 368 |
264319_at |
AT1G04110
|
AtSDD1, SDD1, STOMATAL DENSITY AND DISTRIBUTION 1 |
-0.493 |
0.205 |
| 369 |
254746_at |
AT4G12980
|
[Auxin-responsive family protein] |
-0.493 |
0.073 |
| 370 |
257772_at |
AT3G23080
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.492 |
-0.054 |
| 371 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.492 |
0.092 |
| 372 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
-0.490 |
0.170 |
| 373 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
-0.488 |
0.152 |
| 374 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
-0.487 |
-0.015 |
| 375 |
250589_at |
AT5G07700
|
AtMYB76, myb domain protein 76, MYB76, myb domain protein 76 |
-0.481 |
0.109 |
| 376 |
262693_at |
AT1G62780
|
unknown |
-0.481 |
-0.068 |
| 377 |
266655_at |
AT2G25880
|
AtAUR2, ataurora2, AUR2, ataurora2 |
-0.481 |
0.077 |
| 378 |
260982_at |
AT1G53520
|
AtFAP3, FAP3, fatty-acid-binding protein 3 |
-0.479 |
-0.023 |
| 379 |
248246_at |
AT5G53200
|
TRY, TRIPTYCHON |
-0.479 |
-0.082 |
| 380 |
258333_at |
AT3G16000
|
MFP1, MAR binding filament-like protein 1 |
-0.477 |
-0.042 |
| 381 |
246462_at |
AT5G16940
|
[carbon-sulfur lyases] |
-0.473 |
0.121 |
| 382 |
251155_at |
AT3G63160
|
OEP6, outer envelope protein 6 |
-0.469 |
-0.090 |
| 383 |
262612_at |
AT1G14150
|
PnsL2, Photosynthetic NDH subcomplex L 2, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.466 |
0.036 |
| 384 |
248151_at |
AT5G54270
|
LHCB3, light-harvesting chlorophyll B-binding protein 3, LHCB3*1 |
-0.465 |
0.014 |
| 385 |
264262_at |
AT1G09200
|
H3.1, histone 3.1 |
-0.464 |
0.058 |
| 386 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
-0.464 |
0.169 |
| 387 |
260266_at |
AT1G68520
|
BBX14, B-box domain protein 14 |
-0.458 |
-0.117 |
| 388 |
267635_at |
AT2G42220
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.456 |
0.035 |
| 389 |
254609_at |
AT4G18970
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.454 |
-0.041 |
| 390 |
248247_at |
AT5G53210
|
SPCH, SPEECHLESS |
-0.450 |
-0.002 |
| 391 |
246002_at |
AT5G20740
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.449 |
0.012 |
| 392 |
246159_at |
AT5G20935
|
CRR42, CHLORORESPIRATORY REDUCTION 42 |
-0.449 |
0.020 |
| 393 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
-0.447 |
0.135 |
| 394 |
264096_at |
AT1G78995
|
unknown |
-0.446 |
-0.041 |
| 395 |
267063_at |
AT2G41120
|
unknown |
-0.446 |
-0.074 |
| 396 |
246920_at |
AT5G25090
|
AtENODL13, ENODL13, early nodulin-like protein 13 |
-0.445 |
0.072 |
| 397 |
258067_at |
AT3G25980
|
AtMAD2, MAD2, MITOTIC ARREST-DEFICIENT 2 |
-0.444 |
0.023 |
| 398 |
256469_at |
AT1G32540
|
LOL1, lsd one like 1 |
-0.443 |
0.065 |
| 399 |
251919_at |
AT3G53800
|
Fes1B, Fes1B |
-0.443 |
-0.111 |
| 400 |
250346_at |
AT5G11950
|
LOG8, LONELY GUY 8 |
-0.443 |
0.096 |
| 401 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.442 |
-0.142 |
| 402 |
261422_at |
AT1G18730
|
NDF6, NDH dependent flow 6, PnsB4, Photosynthetic NDH subcomplex B 4 |
-0.441 |
-0.078 |
| 403 |
256015_at |
AT1G19150
|
LHCA2*1, Lhca6, photosystem I light harvesting complex gene 6 |
-0.438 |
-0.049 |
| 404 |
265085_at |
AT1G03780
|
AtTPX2, TPX2, targeting protein for XKLP2 |
-0.436 |
0.036 |
| 405 |
263264_at |
AT2G38810
|
HTA8, histone H2A 8 |
-0.434 |
0.008 |
| 406 |
258920_at |
AT3G10520
|
AHB2, haemoglobin 2, ARATH GLB2, ATGLB2, ARABIDOPSIS HEMOGLOBIN 2, GLB2, HEMOGLOBIN 2, HB2, haemoglobin 2, NSHB2, NON-SYMBIOTIC HAEMOGLOBIN 2 |
-0.431 |
0.087 |
| 407 |
253244_at |
AT4G34580
|
AtSFH1, COW1, CAN OF WORMS1, SRH1, SHORT ROOT HAIR 1 |
-0.431 |
0.015 |
| 408 |
254638_at |
AT4G18740
|
[Rho termination factor] |
-0.430 |
-0.100 |
| 409 |
247962_at |
AT5G56580
|
ANQ1, ARABIDOPSIS NQK1, ATMKK6, ARABIDOPSIS THALIANA MAP KINASE KINASE 6, MKK6, MAP kinase kinase 6 |
-0.430 |
0.037 |
| 410 |
254251_at |
AT4G23300
|
CRK22, cysteine-rich RLK (RECEPTOR-like protein kinase) 22 |
-0.427 |
0.108 |
| 411 |
254398_at |
AT4G21280
|
PSBQ, PHOTOSYSTEM II SUBUNIT Q, PSBQ-1, PHOTOSYSTEM II SUBUNIT Q-1, PSBQA, photosystem II subunit QA |
-0.427 |
0.007 |
| 412 |
251218_at |
AT3G62410
|
CP12, CP12 DOMAIN-CONTAINING PROTEIN 1, CP12-2, CP12 domain-containing protein 2 |
-0.426 |
-0.070 |
| 413 |
262313_at |
AT1G70900
|
unknown |
-0.425 |
0.038 |
| 414 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.424 |
0.082 |
| 415 |
263452_at |
AT2G22190
|
TPPE, trehalose-6-phosphate phosphatase E |
-0.424 |
0.043 |
| 416 |
266551_at |
AT2G35260
|
unknown |
-0.423 |
0.010 |
| 417 |
255773_at |
AT1G18590
|
ATSOT17, SULFOTRANSFERASE 17, ATST5C, ARABIDOPSIS SULFOTRANSFERASE 5C, SOT17, sulfotransferase 17 |
-0.423 |
0.124 |
| 418 |
252149_at |
AT3G51290
|
APSR1, Altered Phosphate Starvation Response 1 |
-0.423 |
-0.126 |
| 419 |
256336_at |
AT1G72030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.420 |
0.023 |
| 420 |
260388_at |
AT1G74070
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.419 |
0.023 |
| 421 |
263674_at |
AT2G04790
|
unknown |
-0.416 |
-0.106 |
| 422 |
257300_at |
AT3G28080
|
UMAMIT47, Usually multiple acids move in and out Transporters 47 |
-0.416 |
-0.072 |
| 423 |
256611_at |
AT3G29270
|
[RING/U-box superfamily protein] |
-0.416 |
-0.065 |
| 424 |
263106_at |
AT2G05160
|
[CCCH-type zinc fingerfamily protein with RNA-binding domain] |
-0.415 |
0.130 |
| 425 |
253044_at |
AT4G37290
|
unknown |
-0.415 |
0.078 |
| 426 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.414 |
0.108 |
| 427 |
257203_at |
AT3G23730
|
XTH16, xyloglucan endotransglucosylase/hydrolase 16 |
-0.414 |
0.030 |
| 428 |
249120_at |
AT5G43750
|
NDH18, NAD(P)H dehydrogenase 18, PnsB5, Photosynthetic NDH subcomplex B 5 |
-0.413 |
0.071 |
| 429 |
258285_at |
AT3G16140
|
PSAH-1, photosystem I subunit H-1 |
-0.413 |
-0.010 |
| 430 |
263495_at |
AT2G42530
|
COR15B, cold regulated 15b |
-0.413 |
0.000 |
| 431 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.410 |
0.047 |
| 432 |
258895_at |
AT3G05600
|
[alpha/beta-Hydrolases superfamily protein] |
-0.410 |
-0.052 |
| 433 |
250434_at |
AT5G10390
|
H3.1, histone 3.1 |
-0.409 |
0.074 |
| 434 |
249472_at |
AT5G39210
|
CRR7, CHLORORESPIRATORY REDUCTION 7 |
-0.409 |
-0.013 |
| 435 |
254545_at |
AT4G19830
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.408 |
0.001 |
| 436 |
265287_at |
AT2G20260
|
PSAE-2, photosystem I subunit E-2 |
-0.408 |
0.003 |
| 437 |
257714_at |
AT3G27360
|
H3.1, histone 3.1 |
-0.407 |
0.064 |
| 438 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
-0.407 |
0.018 |
| 439 |
255460_at |
AT4G02800
|
unknown |
-0.407 |
0.055 |
| 440 |
261981_at |
AT1G33811
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.406 |
-0.052 |
| 441 |
260540_at |
AT2G43500
|
NLP8, NIN-like protein 8 |
-0.405 |
0.036 |
| 442 |
266395_at |
AT2G43100
|
ATLEUD1, IPMI2, isopropylmalate isomerase 2 |
-0.405 |
0.009 |
| 443 |
252199_at |
AT3G50270
|
[HXXXD-type acyl-transferase family protein] |
-0.404 |
-0.143 |
| 444 |
255457_at |
AT4G02770
|
PSAD-1, photosystem I subunit D-1 |
-0.404 |
0.032 |
| 445 |
266979_at |
AT2G39470
|
PnsL1, Photosynthetic NDH subcomplex L 1, PPL2, PsbP-like protein 2 |
-0.402 |
0.004 |
| 446 |
251113_at |
AT5G01370
|
ACI1, ALC-interacting protein 1, TRM29, TON1 Recruiting Motif 29 |
-0.401 |
-0.029 |
| 447 |
258707_at |
AT3G09480
|
[Histone superfamily protein] |
-0.401 |
-0.004 |
| 448 |
252876_at |
AT4G39970
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.401 |
0.031 |
| 449 |
254862_at |
AT4G12030
|
BASS5, BILE ACID:SODIUM SYMPORTER FAMILY PROTEIN 5, BAT5, bile acid transporter 5 |
-0.401 |
0.043 |
| 450 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
-0.399 |
0.451 |
| 451 |
250054_at |
AT5G17860
|
CAX7, calcium exchanger 7 |
-0.399 |
0.080 |
| 452 |
259840_at |
AT1G52230
|
PSAH-2, PHOTOSYSTEM I SUBUNIT H-2, PSAH2, photosystem I subunit H2, PSI-H |
-0.398 |
0.021 |
| 453 |
261247_at |
AT1G20070
|
unknown |
-0.398 |
-0.045 |
| 454 |
246997_at |
AT5G67390
|
unknown |
-0.398 |
0.063 |
| 455 |
265097_at |
AT1G04020
|
ATBARD1, BARD1, breast cancer associated RING 1, ROW1 |
-0.398 |
0.084 |
| 456 |
254783_at |
AT4G12830
|
[alpha/beta-Hydrolases superfamily protein] |
-0.396 |
0.066 |
| 457 |
259707_at |
AT1G77490
|
TAPX, thylakoidal ascorbate peroxidase |
-0.396 |
0.019 |
| 458 |
259768_at |
AT1G29390
|
COR314-TM2, cold regulated 314 thylakoid membrane 2, COR413IM2, COLD REGULATED 314 INNER MEMBRANE 2 |
-0.395 |
-0.006 |
| 459 |
253337_at |
AT4G33470
|
ATHDA14, HDA14, histone deacetylase 14 |
-0.395 |
0.072 |
| 460 |
255265_at |
AT4G05190
|
ATK5, kinesin 5 |
-0.395 |
0.011 |
| 461 |
267637_at |
AT2G42190
|
unknown |
-0.395 |
-0.048 |
| 462 |
251885_at |
AT3G54050
|
HCEF1, high cyclic electron flow 1 |
-0.394 |
0.026 |
| 463 |
259658_at |
AT1G55370
|
NDF5, NDH-dependent cyclic electron flow 5 |
-0.393 |
-0.225 |
| 464 |
246540_at |
AT5G15600
|
SP1L4, SPIRAL1-like4 |
-0.393 |
0.109 |
| 465 |
262543_at |
AT1G34245
|
EPF2, EPIDERMAL PATTERNING FACTOR 2 |
-0.392 |
0.692 |
| 466 |
262412_at |
AT1G34760
|
GF14 OMICRON, GRF11, general regulatory factor 11, RHS5, ROOT HAIR SPECIFIC 5 |
-0.391 |
0.121 |
| 467 |
255284_at |
AT4G04610
|
APR, APR1, APS reductase 1, ATAPR1, PRH19, PAPS REDUCTASE HOMOLOG 19 |
-0.391 |
0.106 |
| 468 |
258326_at |
AT3G22760
|
SOL1 |
-0.391 |
0.054 |
| 469 |
249710_at |
AT5G35630
|
ATGSL1, GLUTAMINE SYNTHETASE LIKE 1, GLN2, GLUTAMINE SYNTHETASE 2, GS2, glutamine synthetase 2 |
-0.390 |
0.003 |
| 470 |
250598_at |
AT5G07690
|
ATMYB29, myb domain protein 29, MYB29, myb domain protein 29, PMG2, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 2 |
-0.389 |
-0.028 |
| 471 |
265547_at |
AT2G28305
|
ATLOG1, LOG1, LONELY GUY 1 |
-0.389 |
-0.180 |
| 472 |
256228_at |
AT1G56190
|
[Phosphoglycerate kinase family protein] |
-0.389 |
0.019 |
| 473 |
250433_at |
AT5G10400
|
H3.1, histone 3.1 |
-0.388 |
0.051 |
| 474 |
250262_at |
AT5G13410
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.388 |
-0.057 |
| 475 |
256125_at |
AT1G18250
|
ATLP-1 |
-0.386 |
0.057 |
| 476 |
248890_at |
AT5G46270
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.386 |
0.023 |
| 477 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
-0.385 |
0.066 |
| 478 |
261075_at |
AT1G07280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.384 |
-0.056 |
| 479 |
248912_at |
AT5G45670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.383 |
-0.093 |
| 480 |
260745_at |
AT1G78370
|
ATGSTU20, glutathione S-transferase TAU 20, GSTU20, glutathione S-transferase TAU 20 |
-0.383 |
0.058 |
| 481 |
265724_at |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
-0.382 |
-0.047 |
| 482 |
260125_at |
AT1G36390
|
[Co-chaperone GrpE family protein] |
-0.382 |
-0.010 |
| 483 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.382 |
-0.043 |
| 484 |
249836_at |
AT5G23420
|
HMGB6, high-mobility group box 6 |
-0.381 |
-0.076 |
| 485 |
266226_at |
AT2G28740
|
HIS4, histone H4 |
-0.380 |
0.025 |
| 486 |
267057_at |
AT2G32500
|
[Stress responsive alpha-beta barrel domain protein] |
-0.380 |
-0.040 |
| 487 |
261016_at |
AT1G26560
|
BGLU40, beta glucosidase 40 |
-0.379 |
0.055 |
| 488 |
248793_at |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
-0.379 |
-0.010 |
| 489 |
260044_at |
AT1G73655
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.379 |
-0.035 |
| 490 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.377 |
0.024 |
| 491 |
249010_at |
AT5G44580
|
unknown |
-0.377 |
0.125 |
| 492 |
248197_at |
AT5G54190
|
PORA, protochlorophyllide oxidoreductase A |
-0.376 |
-0.025 |
| 493 |
259912_at |
AT1G72670
|
iqd8, IQ-domain 8 |
-0.376 |
0.071 |
| 494 |
250633_at |
AT5G07460
|
ATMSRA2, ARABIDOPSIS THALIANA METHIONINE SULFOXIDE REDUCTASE 2, PMSR2, peptidemethionine sulfoxide reductase 2 |
-0.374 |
0.070 |
| 495 |
261248_at |
AT1G20030
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.373 |
0.010 |
| 496 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
-0.373 |
0.178 |
| 497 |
267569_at |
AT2G30790
|
PSBP-2, photosystem II subunit P-2 |
-0.372 |
0.101 |
| 498 |
251755_at |
AT3G55790
|
unknown |
-0.369 |
0.033 |
| 499 |
248687_at |
AT5G48300
|
ADG1, ADP glucose pyrophosphorylase 1, APS1, ADP-GLUCOSE PYROPHOSPHORYLASE SMALL SUBUNIT 1 |
-0.369 |
0.033 |
| 500 |
254150_at |
AT4G24350
|
[Phosphorylase superfamily protein] |
-0.369 |
0.126 |
| 501 |
254137_at |
AT4G24930
|
[thylakoid lumenal 17.9 kDa protein, chloroplast] |
-0.368 |
0.015 |
| 502 |
263607_at |
AT2G16270
|
unknown |
-0.368 |
0.032 |
| 503 |
248963_at |
AT5G45700
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.368 |
-0.017 |
| 504 |
265032_at |
AT1G61580
|
ARP2, ARABIDOPSIS RIBOSOMAL PROTEIN 2, RPL3B, R-protein L3 B |
-0.368 |
-0.005 |
| 505 |
261788_at |
AT1G15980
|
NDF1, NDH-dependent cyclic electron flow 1, NDH48, NAD(P)H DEHYDROGENASE SUBUNIT 48, PnsB1, Photosynthetic NDH subcomplex B 1 |
-0.367 |
-0.001 |
| 506 |
249694_at |
AT5G35790
|
G6PD1, glucose-6-phosphate dehydrogenase 1 |
-0.367 |
-0.057 |
| 507 |
254755_at |
AT4G13220
|
unknown |
-0.367 |
-0.025 |
| 508 |
247606_at |
AT5G61000
|
ATRPA70D, RPA70D |
-0.365 |
0.044 |
| 509 |
264738_at |
AT1G62250
|
unknown |
-0.364 |
0.019 |
| 510 |
262290_at |
AT1G70985
|
[hydroxyproline-rich glycoprotein family protein] |
-0.364 |
0.131 |
| 511 |
263287_at |
AT2G36145
|
unknown |
-0.364 |
-0.018 |
| 512 |
258607_at |
AT3G02730
|
ATF1, TRXF1, thioredoxin F-type 1 |
-0.363 |
0.062 |
| 513 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
-0.362 |
0.011 |
| 514 |
265960_at |
AT2G37470
|
[Histone superfamily protein] |
-0.361 |
0.076 |
| 515 |
256926_at |
AT3G22540
|
unknown |
-0.361 |
0.147 |
| 516 |
256979_at |
AT3G21055
|
PSBTN, photosystem II subunit T |
-0.360 |
-0.005 |
| 517 |
252366_at |
AT3G48420
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.360 |
0.033 |
| 518 |
267402_at |
AT2G26180
|
IQD6, IQ-domain 6 |
-0.359 |
0.011 |
| 519 |
251846_at |
AT3G54560
|
HTA11, histone H2A 11 |
-0.358 |
0.048 |
| 520 |
255013_at |
AT4G10000
|
[Thioredoxin family protein] |
-0.358 |
0.057 |
| 521 |
249355_at |
AT5G40500
|
unknown |
-0.357 |
0.046 |
| 522 |
262669_at |
AT1G62850
|
[Class I peptide chain release factor] |
-0.357 |
-0.081 |
| 523 |
253283_at |
AT4G34090
|
unknown |
-0.357 |
-0.022 |
| 524 |
253480_at |
AT4G31840
|
AtENODL15, ENODL15, early nodulin-like protein 15 |
-0.356 |
0.010 |
| 525 |
251814_at |
AT3G54890
|
LHCA1, photosystem I light harvesting complex gene 1 |
-0.355 |
0.004 |
| 526 |
257021_at |
AT3G19710
|
BCAT4, branched-chain aminotransferase4 |
-0.355 |
0.049 |
| 527 |
248888_at |
AT5G46240
|
AtKAT1, KAT1, potassium channel in Arabidopsis thaliana 1 |
-0.355 |
0.043 |
| 528 |
253860_at |
AT4G27700
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.354 |
0.001 |
| 529 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
-0.350 |
0.080 |
| 530 |
267220_at |
AT2G02500
|
ATMEPCT, ISPD, MCT, 2-C-METHYL-D-ERYTHRITOL 4-PHOSPHATE CYTIDYLTRANSFERASE |
-0.350 |
0.004 |
| 531 |
266222_at |
AT2G28780
|
unknown |
-0.349 |
0.010 |
| 532 |
246557_at |
AT5G15510
|
[TPX2 (targeting protein for Xklp2) protein family] |
-0.348 |
0.034 |
| 533 |
252089_at |
AT3G52110
|
unknown |
-0.347 |
0.060 |
| 534 |
248624_at |
AT5G48790
|
unknown |
-0.346 |
-0.046 |
| 535 |
250228_at |
AT5G13840
|
CCS52B, cell cycle switch protein 52 B, FZR3, FIZZY-related 3 |
-0.346 |
0.015 |
| 536 |
252661_at |
AT3G44450
|
unknown |
-0.346 |
0.045 |
| 537 |
256400_at |
AT3G06140
|
LUL4, LOG2-LIKE UBIQUITIN LIGASE4 |
-0.345 |
0.068 |
| 538 |
254125_at |
AT4G24670
|
TAR2, tryptophan aminotransferase related 2 |
-0.345 |
-0.016 |
| 539 |
258530_at |
AT3G06840
|
unknown |
-0.345 |
0.142 |
| 540 |
252574_at |
AT3G45430
|
LecRK-I.5, L-type lectin receptor kinase I.5 |
-0.345 |
0.092 |
| 541 |
265349_at |
AT2G22610
|
[Di-glucose binding protein with Kinesin motor domain] |
-0.344 |
0.039 |
| 542 |
262236_at |
AT1G48330
|
unknown |
-0.343 |
-0.054 |
| 543 |
250255_at |
AT5G13730
|
SIG4, sigma factor 4, SIGD |
-0.343 |
-0.082 |
| 544 |
263142_at |
AT1G65230
|
[Uncharacterized conserved protein (DUF2358)] |
-0.343 |
0.000 |
| 545 |
250453_at |
AT5G10620
|
[methyltransferases] |
-0.342 |
0.032 |
| 546 |
245082_at |
AT2G23270
|
unknown |
-0.342 |
0.173 |
| 547 |
245213_at |
AT1G44575
|
CP22, NPQ4, NONPHOTOCHEMICAL QUENCHING 4, PSBS, PHOTOSYSTEM II SUBUNIT S |
-0.342 |
0.009 |
| 548 |
246315_at |
AT3G56870
|
unknown |
-0.341 |
0.025 |
| 549 |
261026_at |
AT1G01240
|
unknown |
-0.341 |
0.035 |
| 550 |
258932_at |
AT3G10150
|
ATPAP16, PAP16, purple acid phosphatase 16 |
-0.340 |
0.108 |
| 551 |
257115_at |
AT3G20150
|
[Kinesin motor family protein] |
-0.339 |
-0.110 |
| 552 |
261801_at |
AT1G30520
|
AAE14, acyl-activating enzyme 14 |
-0.339 |
0.027 |
| 553 |
267118_at |
AT2G32590
|
EMB2795, EMBRYO DEFECTIVE 2795 |
-0.338 |
0.071 |
| 554 |
251067_at |
AT5G01910
|
unknown |
-0.338 |
0.029 |
| 555 |
261346_at |
AT1G79720
|
[Eukaryotic aspartyl protease family protein] |
-0.337 |
0.031 |
| 556 |
257801_at |
AT3G18750
|
ATWNK6, ARABIDOPSIS THALIANA WITH NO K 6, WNK6, with no lysine (K) kinase 6, ZIK5 |
-0.337 |
0.115 |
| 557 |
261218_at |
AT1G20020
|
ATLFNR2, LEAF FNR 2, FNR2, ferredoxin-NADP(+)-oxidoreductase 2, LFNR2, leaf-type chloroplast-targeted FNR 2 |
-0.336 |
0.028 |
| 558 |
245242_at |
AT1G44446
|
ATCAO, ARABIDOPSIS THALIANA CHLOROPHYLL A OXYGENASE, CAO, CHLOROPHYLL A OXYGENASE, CH1, CHLORINA 1 |
-0.336 |
0.024 |
| 559 |
253343_at |
AT4G33540
|
[metallo-beta-lactamase family protein] |
-0.336 |
0.062 |
| 560 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
-0.335 |
0.006 |
| 561 |
267359_at |
AT2G40020
|
[Nucleolar histone methyltransferase-related protein] |
-0.335 |
0.044 |
| 562 |
247651_at |
AT5G59870
|
HTA6, histone H2A 6 |
-0.333 |
0.041 |
| 563 |
256441_at |
AT3G10940
|
LSF2, LIKE SEX4 2 |
-0.332 |
0.049 |
| 564 |
264845_at |
AT1G03680
|
ATHM1, thioredoxin M-type 1, ATM1, ARABIDOPSIS THIOREDOXIN M-TYPE 1, THM1, thioredoxin M-type 1, TRX-M1, THIOREDOXIN M-TYPE 1 |
-0.332 |
0.023 |
| 565 |
252114_at |
AT3G51450
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.332 |
0.006 |
| 566 |
251485_at |
AT3G59550
|
ATRAD21.2, SISTER CHROMATID COHESION 1 PROTEIN 3, ATSYN3, SYN3 |
-0.331 |
0.070 |
| 567 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.330 |
-0.141 |
| 568 |
262109_at |
AT1G02730
|
ATCSLD5, cellulose synthase-like D5, CSLD5, CELLULOSE SYNTHASE LIKE D5, CSLD5, cellulose synthase-like D5, SOS6, SALT OVERLY SENSITIVE 6 |
-0.330 |
0.029 |
| 569 |
252148_at |
AT3G51280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.330 |
0.036 |
| 570 |
252001_at |
AT3G52750
|
FTSZ2-2 |
-0.329 |
0.059 |
| 571 |
258098_at |
AT3G23670
|
KINESIN-12B, PAKRP1L |
-0.329 |
0.118 |
| 572 |
260685_at |
AT1G17650
|
AtGLYR2, GLYR2, glyoxylate reductase 2, GR2, GLYOXYLATE REDUCTASE 2 |
-0.329 |
0.032 |
| 573 |
250561_at |
AT5G08030
|
AtGDPD6, GDPD6, glycerophosphodiester phosphodiesterase 6 |
-0.329 |
0.054 |
| 574 |
255433_at |
AT4G03210
|
XTH9, xyloglucan endotransglucosylase/hydrolase 9 |
-0.327 |
0.021 |
| 575 |
247400_at |
AT5G62840
|
[Phosphoglycerate mutase family protein] |
-0.326 |
0.091 |
| 576 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
-0.326 |
-0.068 |
| 577 |
250613_at |
AT5G07240
|
IQD24, IQ-domain 24 |
-0.326 |
0.095 |
| 578 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
-0.326 |
-0.082 |
| 579 |
249927_at |
AT5G19220
|
ADG2, ADP GLUCOSE PYROPHOSPHORYLASE 2, APL1, ADP glucose pyrophosphorylase large subunit 1 |
-0.325 |
-0.031 |
| 580 |
247425_at |
AT5G62550
|
unknown |
-0.325 |
0.049 |
| 581 |
267622_at |
AT2G39690
|
unknown |
-0.325 |
0.008 |
| 582 |
259838_at |
AT1G52220
|
CURT1C, CURVATURE THYLAKOID 1C |
-0.325 |
0.020 |
| 583 |
265819_at |
AT2G17972
|
unknown |
-0.325 |
0.056 |