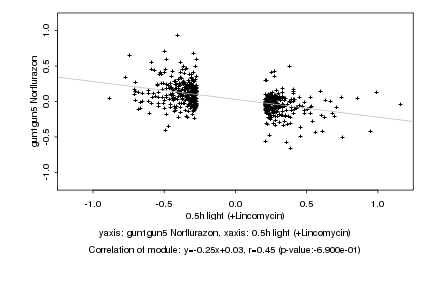
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
0.5h light (+Lincomycin) |
Log2 signal ratio
gun1gun5 Norflurazon |
| 1 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
1.156 |
-0.030 |
| 2 |
265611_at |
AT2G25510
|
unknown |
0.984 |
0.140 |
| 3 |
263161_at |
AT1G54020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.946 |
-0.417 |
| 4 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
0.852 |
0.047 |
| 5 |
256459_at |
AT1G36180
|
ACC2, acetyl-CoA carboxylase 2 |
0.748 |
-0.499 |
| 6 |
257592_at |
AT3G24982
|
ATRLP40, receptor like protein 40, RLP40, receptor like protein 40 |
0.739 |
0.057 |
| 7 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
0.706 |
-0.082 |
| 8 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
0.692 |
-0.209 |
| 9 |
263515_at |
AT2G21640
|
unknown |
0.681 |
-0.160 |
| 10 |
259561_at |
AT1G21250
|
AtWAK1, PRO25, WAK1, cell wall-associated kinase 1 |
0.661 |
0.005 |
| 11 |
259925_at |
AT1G75040
|
PR-5, PR5, pathogenesis-related gene 5 |
0.627 |
0.018 |
| 12 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
0.625 |
-0.215 |
| 13 |
264635_at |
AT1G65500
|
unknown |
0.610 |
-0.412 |
| 14 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.602 |
-0.196 |
| 15 |
250435_at |
AT5G10380
|
ATRING1, RING1 |
0.595 |
0.148 |
| 16 |
267076_at |
AT2G41090
|
[Calcium-binding EF-hand family protein] |
0.556 |
-0.424 |
| 17 |
259553_x_at |
AT1G21310
|
ATEXT3, extensin 3, EXT3, extensin 3, RSH, ROOT-SHOOT-HYPOCOTYL DEFECTIVE |
0.536 |
-0.278 |
| 18 |
251842_at |
AT3G54580
|
[Proline-rich extensin-like family protein] |
0.528 |
-0.058 |
| 19 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
0.521 |
0.059 |
| 20 |
257735_at |
AT3G27400
|
[Pectin lyase-like superfamily protein] |
0.520 |
-0.162 |
| 21 |
249895_at |
AT5G22500
|
FAR1, fatty acid reductase 1 |
0.520 |
-0.073 |
| 22 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
0.499 |
-0.102 |
| 23 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.483 |
-0.124 |
| 24 |
251843_x_at |
AT3G54590
|
ATHRGP1, hydroxyproline-rich glycoprotein, HRGP1, hydroxyproline-rich glycoprotein |
0.481 |
-0.010 |
| 25 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
0.478 |
-0.161 |
| 26 |
254635_at |
AT4G18670
|
[Leucine-rich repeat (LRR) family protein] |
0.469 |
-0.060 |
| 27 |
254832_at |
AT4G12490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.453 |
-0.489 |
| 28 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
0.453 |
-0.359 |
| 29 |
257667_at |
AT3G20440
|
BE1, BRANCHING ENZYME 1, EMB2729, EMBRYO DEFECTIVE 2729 |
0.448 |
-0.043 |
| 30 |
264886_at |
AT1G61120
|
GES, GERANYLLINALOOL SYNTHASE, TPS04, terpene synthase 04, TPS4, TERPENE SYNTHASE 4 |
0.447 |
0.061 |
| 31 |
254770_at |
AT4G13340
|
LRX3, leucine-rich repeat/extensin 3 |
0.434 |
-0.075 |
| 32 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.426 |
-0.074 |
| 33 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
0.424 |
-0.098 |
| 34 |
266352_at |
AT2G01610
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.413 |
-0.102 |
| 35 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
0.408 |
0.036 |
| 36 |
265536_at |
AT2G15880
|
[Leucine-rich repeat (LRR) family protein] |
0.407 |
0.150 |
| 37 |
258063_at |
AT3G14620
|
CYP72A8, cytochrome P450, family 72, subfamily A, polypeptide 8 |
0.405 |
0.125 |
| 38 |
247615_at |
AT5G60250
|
[zinc finger (C3HC4-type RING finger) family protein] |
0.405 |
-0.167 |
| 39 |
259802_at |
AT1G72260
|
THI2.1, thionin 2.1, THI2.1.1 |
0.405 |
0.030 |
| 40 |
258961_at |
AT3G10580
|
[Homeodomain-like superfamily protein] |
0.405 |
0.012 |
| 41 |
257333_at |
ATMG01360
|
COX1, cytochrome oxidase |
0.402 |
0.176 |
| 42 |
256188_at |
AT1G30160
|
unknown |
0.392 |
-0.071 |
| 43 |
262414_at |
AT1G49430
|
LACS2, long-chain acyl-CoA synthetase 2, LRD2, LATERAL ROOT DEVELOPMENT 2 |
0.391 |
-0.124 |
| 44 |
253763_at |
AT4G28850
|
ATXTH26, XTH26, xyloglucan endotransglucosylase/hydrolase 26 |
0.388 |
0.041 |
| 45 |
266503_at |
AT2G47780
|
[Rubber elongation factor protein (REF)] |
0.386 |
-0.307 |
| 46 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
0.386 |
-0.007 |
| 47 |
249784_at |
AT5G24280
|
GMI1, GAMMA-IRRADIATION AND MITOMYCIN C INDUCED 1 |
0.385 |
-0.216 |
| 48 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
0.383 |
-0.652 |
| 49 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
0.376 |
0.494 |
| 50 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
0.374 |
0.004 |
| 51 |
253207_at |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
0.366 |
-0.201 |
| 52 |
249777_at |
AT5G24210
|
[alpha/beta-Hydrolases superfamily protein] |
0.365 |
-0.038 |
| 53 |
250155_at |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
0.364 |
-0.311 |
| 54 |
247880_at |
AT5G57780
|
P1R1, P1R1 |
0.356 |
-0.124 |
| 55 |
248509_at |
AT5G50335
|
unknown |
0.354 |
-0.574 |
| 56 |
253051_at |
AT4G37490
|
CYC1, CYCLIN 1, CYCB1, CYCB1;1, CYCLIN B1;1 |
0.350 |
-0.204 |
| 57 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
0.349 |
-0.209 |
| 58 |
254287_at |
AT4G22960
|
unknown |
0.348 |
-0.189 |
| 59 |
258523_at |
AT3G06830
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.345 |
0.049 |
| 60 |
251826_at |
AT3G55110
|
ABCG18, ATP-binding cassette G18 |
0.343 |
-0.067 |
| 61 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
0.342 |
-0.066 |
| 62 |
249476_at |
AT5G38910
|
[RmlC-like cupins superfamily protein] |
0.342 |
0.021 |
| 63 |
264868_at |
AT1G24095
|
[Putative thiol-disulphide oxidoreductase DCC] |
0.341 |
-0.061 |
| 64 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.340 |
0.053 |
| 65 |
245196_at |
AT1G67750
|
[Pectate lyase family protein] |
0.339 |
-0.278 |
| 66 |
248619_at |
AT5G49630
|
AAP6, amino acid permease 6 |
0.337 |
-0.048 |
| 67 |
252437_at |
AT3G47380
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.333 |
-0.329 |
| 68 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
0.333 |
-0.054 |
| 69 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.330 |
0.062 |
| 70 |
265083_at |
AT1G03820
|
unknown |
0.330 |
-0.138 |
| 71 |
246850_at |
AT5G26860
|
LON1, lon protease 1, LON_ARA_ARA |
0.330 |
-0.040 |
| 72 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
0.330 |
0.043 |
| 73 |
258920_at |
AT3G10520
|
AHB2, haemoglobin 2, ARATH GLB2, ATGLB2, ARABIDOPSIS HEMOGLOBIN 2, GLB2, HEMOGLOBIN 2, HB2, haemoglobin 2, NSHB2, NON-SYMBIOTIC HAEMOGLOBIN 2 |
0.329 |
-0.073 |
| 74 |
252888_at |
AT4G39210
|
APL3 |
0.328 |
-0.069 |
| 75 |
252359_at |
AT3G48440
|
[Zinc finger C-x8-C-x5-C-x3-H type family protein] |
0.327 |
-0.031 |
| 76 |
256489_at |
AT1G31550
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.327 |
0.184 |
| 77 |
256825_at |
AT3G22120
|
CWLP, cell wall-plasma membrane linker protein |
0.326 |
-0.078 |
| 78 |
260158_at |
AT1G79910
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.325 |
0.076 |
| 79 |
247812_at |
AT5G58390
|
[Peroxidase superfamily protein] |
0.324 |
-0.196 |
| 80 |
252019_at |
AT3G53040
|
[late embryogenesis abundant protein, putative / LEA protein, putative] |
0.324 |
-0.039 |
| 81 |
263411_at |
AT2G28710
|
[C2H2-type zinc finger family protein] |
0.324 |
0.128 |
| 82 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
0.317 |
-0.189 |
| 83 |
252281_at |
AT3G49320
|
[Metal-dependent protein hydrolase] |
0.314 |
0.016 |
| 84 |
252238_at |
AT3G49960
|
[Peroxidase superfamily protein] |
0.314 |
-0.063 |
| 85 |
262290_at |
AT1G70985
|
[hydroxyproline-rich glycoprotein family protein] |
0.312 |
0.048 |
| 86 |
245875_at |
AT1G26240
|
[Proline-rich extensin-like family protein] |
0.309 |
-0.169 |
| 87 |
247463_at |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
0.308 |
-0.169 |
| 88 |
252799_at |
AT3G42320
|
unknown |
0.308 |
0.038 |
| 89 |
248961_at |
AT5G45650
|
[subtilase family protein] |
0.305 |
-0.043 |
| 90 |
258675_at |
AT3G08770
|
LTP6, lipid transfer protein 6 |
0.303 |
-0.069 |
| 91 |
256497_at |
AT1G31580
|
CXC750, ECS1 |
0.303 |
-0.287 |
| 92 |
261192_at |
AT1G32870
|
ANAC013, Arabidopsis NAC domain containing protein 13, ANAC13, NAC domain protein 13, NAC13, NAC domain protein 13 |
0.303 |
-0.046 |
| 93 |
260359_at |
AT1G69210
|
[Uncharacterised protein family UPF0090] |
0.303 |
0.074 |
| 94 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
0.302 |
-0.094 |
| 95 |
260181_at |
AT1G70710
|
ATGH9B1, glycosyl hydrolase 9B1, CEL1, CELLULASE 1, GH9B1, glycosyl hydrolase 9B1 |
0.300 |
-0.051 |
| 96 |
264195_at |
AT1G22690
|
[Gibberellin-regulated family protein] |
0.299 |
-0.098 |
| 97 |
254255_at |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
0.298 |
-0.062 |
| 98 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
0.298 |
0.116 |
| 99 |
255516_at |
AT4G02270
|
RHS13, root hair specific 13 |
0.297 |
0.043 |
| 100 |
261037_at |
AT1G17420
|
ATLOX3, Arabidopsis thaliana lipoxygenase 3, LOX3, lipoxygenase 3 |
0.296 |
0.037 |
| 101 |
263899_at |
AT2G21710
|
EMB2219, embryo defective 2219 |
0.295 |
0.080 |
| 102 |
257525_at |
AT3G04900
|
[Heavy metal transport/detoxification superfamily protein ] |
0.294 |
0.050 |
| 103 |
257474_at |
AT1G80850
|
[DNA glycosylase superfamily protein] |
0.294 |
-0.026 |
| 104 |
263998_at |
AT2G22510
|
[hydroxyproline-rich glycoprotein family protein] |
0.291 |
-0.198 |
| 105 |
262508_at |
AT1G11300
|
[protein serine/threonine kinases] |
0.291 |
-0.068 |
| 106 |
247377_at |
AT5G63180
|
[Pectin lyase-like superfamily protein] |
0.290 |
-0.136 |
| 107 |
267121_at |
AT2G23540
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.290 |
-0.198 |
| 108 |
266511_at |
AT2G47680
|
[zinc finger (CCCH type) helicase family protein] |
0.287 |
-0.025 |
| 109 |
266154_at |
AT2G12190
|
[Cytochrome P450 superfamily protein] |
0.287 |
0.161 |
| 110 |
259609_at |
AT1G52410
|
AtTSA1, TSA1, TSK-associating protein 1 |
0.287 |
-0.008 |
| 111 |
252740_at |
AT3G43270
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.286 |
-0.032 |
| 112 |
249576_at |
AT5G37690
|
[SGNH hydrolase-type esterase superfamily protein] |
0.286 |
-0.082 |
| 113 |
255628_at |
AT4G00850
|
GIF3, GRF1-interacting factor 3 |
0.285 |
-0.019 |
| 114 |
255973_at |
AT3G32400
|
[Actin-binding FH2/DRF autoregulatory protein] |
0.285 |
-0.071 |
| 115 |
249425_at |
AT5G39790
|
5'-AMP-activated protein kinase-related |
0.285 |
0.082 |
| 116 |
249061_at |
AT5G44550
|
[Uncharacterised protein family (UPF0497)] |
0.283 |
-0.150 |
| 117 |
248605_at |
AT5G49410
|
unknown |
0.283 |
0.057 |
| 118 |
253650_at |
AT4G30020
|
[PA-domain containing subtilase family protein] |
0.282 |
-0.138 |
| 119 |
265155_at |
AT1G30990
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.282 |
0.045 |
| 120 |
253301_at |
AT4G33720
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.279 |
-0.054 |
| 121 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
0.278 |
0.098 |
| 122 |
248333_at |
AT5G52390
|
[PAR1 protein] |
0.278 |
-0.012 |
| 123 |
254189_at |
AT4G24000
|
ATCSLG2, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G2, CSLG2, cellulose synthase like G2 |
0.278 |
0.064 |
| 124 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
0.277 |
-0.209 |
| 125 |
246516_at |
AT5G15740
|
[O-fucosyltransferase family protein] |
0.277 |
-0.077 |
| 126 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
0.277 |
-0.325 |
| 127 |
247762_at |
AT5G59170
|
[Proline-rich extensin-like family protein] |
0.276 |
0.049 |
| 128 |
252653_at |
AT3G44730
|
AtKIN14h, ATKP1, ARABIDOPSIS KINESIN-LIKE PROTEIN 1, KP1, kinesin-like protein 1 |
0.276 |
0.030 |
| 129 |
251250_at |
AT3G62180
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.276 |
0.018 |
| 130 |
261502_at |
AT1G14440
|
AtHB31, homeobox protein 31, FTM2, FLORAL TRANSITION AT THE MERISTEM2, HB31, homeobox protein 31, ZHD4, ZINC FINGER HOMEODOMAIN 4 |
0.275 |
-0.255 |
| 131 |
252594_at |
AT3G45680
|
[Major facilitator superfamily protein] |
0.275 |
0.003 |
| 132 |
267083_at |
AT2G41100
|
ATCAL4, ARABIDOPSIS THALIANA CALMODULIN LIKE 4, TCH3, TOUCH 3 |
0.275 |
-0.016 |
| 133 |
260642_at |
AT1G53260
|
[LOCATED IN: endomembrane system] |
0.274 |
0.055 |
| 134 |
263498_at |
AT2G42610
|
LSH10, LIGHT SENSITIVE HYPOCOTYLS 10 |
0.272 |
0.076 |
| 135 |
244983_at |
ATCG00790
|
RPL16, ribosomal protein L16 |
0.272 |
-0.054 |
| 136 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
0.272 |
0.356 |
| 137 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.270 |
0.425 |
| 138 |
265121_at |
AT1G62560
|
FMO GS-OX3, flavin-monooxygenase glucosinolate S-oxygenase 3 |
0.270 |
0.057 |
| 139 |
265122_at |
AT1G62540
|
FMO GS-OX2, flavin-monooxygenase glucosinolate S-oxygenase 2 |
0.269 |
0.024 |
| 140 |
245076_at |
AT2G23170
|
GH3.3 |
0.268 |
0.093 |
| 141 |
261339_at |
AT1G35710
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.268 |
0.018 |
| 142 |
246986_at |
AT5G67280
|
RLK, receptor-like kinase |
0.268 |
-0.102 |
| 143 |
260110_at |
AT1G63350
|
[Disease resistance protein (CC-NBS-LRR class) family] |
0.266 |
0.047 |
| 144 |
250517_at |
AT5G08260
|
scpl35, serine carboxypeptidase-like 35 |
0.266 |
-0.138 |
| 145 |
246212_at |
AT4G36930
|
SPT, SPATULA |
0.266 |
-0.091 |
| 146 |
250045_at |
AT5G17700
|
[MATE efflux family protein] |
0.266 |
-0.129 |
| 147 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
0.266 |
-0.162 |
| 148 |
248613_at |
AT5G49555
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
0.264 |
-0.115 |
| 149 |
258342_at |
AT3G22800
|
[Leucine-rich repeat (LRR) family protein] |
0.264 |
-0.185 |
| 150 |
260410_at |
AT1G69870
|
AtNPF2.13, NPF2.13, NRT1/ PTR family 2.13, NRT1.7, nitrate transporter 1.7 |
0.263 |
0.029 |
| 151 |
256125_at |
AT1G18250
|
ATLP-1 |
0.262 |
-0.145 |
| 152 |
253659_at |
AT4G30150
|
unknown |
0.262 |
-0.085 |
| 153 |
247252_at |
AT5G64770
|
CLEL 9, CLE-like 9, GLV2, GOLVEN 2, RGF9, root meristem growth factor 9 |
0.261 |
-0.301 |
| 154 |
265926_at |
AT2G18600
|
[Ubiquitin-conjugating enzyme family protein] |
0.260 |
0.089 |
| 155 |
253024_at |
AT4G38080
|
[hydroxyproline-rich glycoprotein family protein] |
0.259 |
-0.143 |
| 156 |
251695_at |
AT3G56590
|
[hydroxyproline-rich glycoprotein family protein] |
0.259 |
0.008 |
| 157 |
257644_at |
AT3G25780
|
AOC3, allene oxide cyclase 3 |
0.257 |
-0.026 |
| 158 |
262750_at |
AT1G28710
|
[Nucleotide-diphospho-sugar transferase family protein] |
0.257 |
-0.096 |
| 159 |
251785_at |
AT3G55130
|
ABCG19, ATP-binding cassette G19, ATWBC19, white-brown complex homolog 19, WBC19, white-brown complex homolog 19 |
0.256 |
0.035 |
| 160 |
256339_at |
AT1G72080
|
unknown |
0.255 |
0.035 |
| 161 |
248152_at |
AT5G54260
|
ATMRE11, ARABIDOPSIS MEIOTIC RECOMBINATION 11, MRE11, MEIOTIC RECOMBINATION 11 |
0.255 |
0.065 |
| 162 |
261242_at |
AT1G32960
|
ATSBT3.3, SBT3.3 |
0.255 |
0.138 |
| 163 |
253682_at |
AT4G29640
|
[Cytidine/deoxycytidylate deaminase family protein] |
0.254 |
0.048 |
| 164 |
251485_at |
AT3G59550
|
ATRAD21.2, SISTER CHROMATID COHESION 1 PROTEIN 3, ATSYN3, SYN3 |
0.254 |
0.012 |
| 165 |
251730_at |
AT3G56330
|
[N2,N2-dimethylguanosine tRNA methyltransferase] |
0.254 |
0.076 |
| 166 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.253 |
0.013 |
| 167 |
245381_at |
AT4G17785
|
MYB39, myb domain protein 39 |
0.253 |
0.013 |
| 168 |
259996_at |
AT1G67910
|
unknown |
0.253 |
-0.220 |
| 169 |
260553_at |
AT2G41800
|
unknown |
0.252 |
-0.014 |
| 170 |
261033_at |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
0.252 |
0.040 |
| 171 |
247612_at |
AT5G60730
|
[Anion-transporting ATPase] |
0.251 |
-0.089 |
| 172 |
267123_at |
AT2G23560
|
ATMES7, ARABIDOPSIS THALIANA METHYL ESTERASE 7, MES7, methyl esterase 7 |
0.251 |
0.076 |
| 173 |
254243_at |
AT4G23210
|
CRK13, cysteine-rich RLK (RECEPTOR-like protein kinase) 13 |
0.250 |
0.075 |
| 174 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
0.249 |
0.416 |
| 175 |
252747_at |
AT3G43320
|
unknown |
0.249 |
0.045 |
| 176 |
250267_at |
AT5G12930
|
unknown |
0.248 |
-0.100 |
| 177 |
246878_at |
AT5G26060
|
[Plant self-incompatibility protein S1 family] |
0.248 |
-0.004 |
| 178 |
260434_at |
AT1G68330
|
unknown |
0.246 |
-0.062 |
| 179 |
266790_at |
AT2G28950
|
ATEXP6, ARABIDOPSIS THALIANA TEXPANSIN 6, ATEXPA6, expansin A6, ATHEXP ALPHA 1.8, EXPA6, expansin A6 |
0.246 |
-0.190 |
| 180 |
263536_at |
AT2G25000
|
ATWRKY60, WRKY60, WRKY DNA-binding protein 60 |
0.246 |
0.065 |
| 181 |
249894_at |
AT5G22580
|
[Stress responsive A/B Barrel Domain] |
0.244 |
-0.246 |
| 182 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
0.243 |
-0.107 |
| 183 |
244957_at |
ATMG00400
|
ORF157 |
0.242 |
0.119 |
| 184 |
264557_at |
AT1G09550
|
[Pectinacetylesterase family protein] |
0.241 |
0.105 |
| 185 |
247352_at |
AT5G63650
|
SNRK2-5, SNRK2.5, SNF1-related protein kinase 2.5, SRK2H, SNF1-RELATED PROTEIN KINASE 2H |
0.240 |
-0.079 |
| 186 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
0.239 |
-0.165 |
| 187 |
256851_at |
AT3G27930
|
unknown |
0.239 |
-0.031 |
| 188 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
0.239 |
-0.173 |
| 189 |
249634_at |
AT5G36840
|
unknown |
0.239 |
0.054 |
| 190 |
256109_at |
AT1G16950
|
unknown |
0.238 |
0.082 |
| 191 |
262829_at |
AT1G14970
|
[O-fucosyltransferase family protein] |
0.238 |
-0.012 |
| 192 |
260666_at |
AT1G19300
|
ATGATL1, GATL1, GALACTURONOSYLTRANSFERASE-LIKE 1, GLZ1, GAOLAOZHUANGREN 1, PARVUS, PARVUS |
0.237 |
-0.116 |
| 193 |
257835_at |
AT3G25180
|
CYP82G1, cytochrome P450, family 82, subfamily G, polypeptide 1 |
0.237 |
0.065 |
| 194 |
258685_at |
AT3G07830
|
PGA3, polygalacturonase 3 |
0.236 |
0.001 |
| 195 |
259640_at |
AT1G52400
|
ATBG1, A. THALIANA BETA-GLUCOSIDASE 1, BGL1, BETA-GLUCOSIDASE HOMOLOG 1, BGLU18, beta glucosidase 18 |
0.236 |
-0.238 |
| 196 |
263034_at |
AT1G24020
|
MLP423, MLP-like protein 423 |
0.235 |
-0.232 |
| 197 |
245087_at |
AT2G39830
|
DAR2, DA1-related protein 2, LRD3, LATERAL ROOT DEVELOPMENT 3 |
0.235 |
-0.002 |
| 198 |
256364_at |
AT1G66550
|
ATWRKY67, WRKY67, WRKY DNA-binding protein 67 |
0.235 |
0.057 |
| 199 |
248895_at |
AT5G46330
|
FLS2, FLAGELLIN-SENSITIVE 2 |
0.234 |
-0.472 |
| 200 |
250719_at |
AT5G06250
|
DPA4, DEVELOPMENT-RELATED PcG TARGET IN THE APEX 4 |
0.233 |
0.051 |
| 201 |
249732_at |
AT5G24420
|
PGL5, 6-phosphogluconolactonase 5 |
0.233 |
0.078 |
| 202 |
257600_at |
AT3G24770
|
CLE41, CLAVATA3/ESR-RELATED 41 |
0.233 |
-0.122 |
| 203 |
257120_at |
AT3G20200
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
0.232 |
-0.029 |
| 204 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
0.232 |
-0.072 |
| 205 |
245688_at |
AT1G28290
|
AGP31, arabinogalactan protein 31 |
0.231 |
-0.222 |
| 206 |
267546_at |
AT2G32680
|
AtRLP23, receptor like protein 23, RLP23, receptor like protein 23 |
0.231 |
0.100 |
| 207 |
250540_at |
AT5G08580
|
[Calcium-binding EF hand family protein] |
0.231 |
-0.062 |
| 208 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.230 |
0.085 |
| 209 |
248870_at |
AT5G46710
|
[PLATZ transcription factor family protein] |
0.230 |
0.001 |
| 210 |
264890_at |
AT1G23180
|
[ARM repeat superfamily protein] |
0.230 |
0.021 |
| 211 |
247958_at |
AT5G57070
|
[hydroxyproline-rich glycoprotein family protein] |
0.230 |
0.031 |
| 212 |
250633_at |
AT5G07460
|
ATMSRA2, ARABIDOPSIS THALIANA METHIONINE SULFOXIDE REDUCTASE 2, PMSR2, peptidemethionine sulfoxide reductase 2 |
0.230 |
-0.046 |
| 213 |
255517_at |
AT4G02290
|
AtGH9B13, glycosyl hydrolase 9B13, GH9B13, glycosyl hydrolase 9B13 |
0.229 |
-0.066 |
| 214 |
246118_at |
AT5G20340
|
BG5, beta-1,3-glucanase 5 |
0.229 |
-0.012 |
| 215 |
251360_at |
AT3G61210
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.229 |
0.005 |
| 216 |
253040_at |
AT4G37800
|
XTH7, xyloglucan endotransglucosylase/hydrolase 7 |
0.228 |
-0.213 |
| 217 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
0.228 |
-0.177 |
| 218 |
265411_at |
AT2G16630
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.228 |
0.078 |
| 219 |
256213_at |
AT1G50990
|
BSK11, brassinosteroid-signaling kinase 11 |
0.228 |
0.018 |
| 220 |
261398_at |
AT1G79610
|
ATNHX6, ARABIDOPSIS NA+/H+ ANTIPORTER 6, NHX6, Na+/H+ antiporter 6 |
0.227 |
0.030 |
| 221 |
244980_at |
ATCG00760
|
RPL36, ribosomal protein L36 |
0.227 |
-0.126 |
| 222 |
263120_at |
AT1G78490
|
CYP708A3, cytochrome P450, family 708, subfamily A, polypeptide 3 |
0.227 |
-0.099 |
| 223 |
259533_at |
AT1G12530
|
MOS9, modifier of snc1 9 |
0.227 |
-0.001 |
| 224 |
257059_at |
AT3G15280
|
unknown |
0.226 |
-0.002 |
| 225 |
256366_at |
AT1G66880
|
[Protein kinase superfamily protein] |
0.226 |
-0.103 |
| 226 |
265895_at |
AT2G15000
|
unknown |
0.226 |
-0.112 |
| 227 |
260234_at |
AT1G74460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.225 |
-0.076 |
| 228 |
253083_at |
AT4G36250
|
ALDH3F1, aldehyde dehydrogenase 3F1 |
0.224 |
-0.105 |
| 229 |
260804_at |
AT1G78410
|
[VQ motif-containing protein] |
0.224 |
-0.016 |
| 230 |
252677_at |
AT3G44320
|
AtNIT3, NITRILASE 3, NIT3, nitrilase 3 |
0.224 |
-0.081 |
| 231 |
257277_at |
AT3G14470
|
[NB-ARC domain-containing disease resistance protein] |
0.224 |
-0.034 |
| 232 |
263738_at |
AT1G60060
|
[Serine/threonine-protein kinase WNK (With No Lysine)-related] |
0.223 |
-0.003 |
| 233 |
265441_at |
AT2G20870
|
[cell wall protein precursor, putative] |
0.223 |
0.041 |
| 234 |
264293_at |
AT1G78770
|
APC6, anaphase promoting complex 6 |
0.223 |
-0.065 |
| 235 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
0.222 |
-0.323 |
| 236 |
251116_at |
AT3G63470
|
scpl40, serine carboxypeptidase-like 40 |
0.222 |
0.051 |
| 237 |
259737_at |
AT1G64400
|
LACS3, long-chain acyl-CoA synthetase 3 |
0.222 |
-0.053 |
| 238 |
245157_at |
AT2G33160
|
NMA, NIMNA (Sanskrit for 'sunken or low) |
0.222 |
0.020 |
| 239 |
259549_at |
AT1G35290
|
ALT1, acyl-lipid thioesterase 1 |
0.222 |
0.031 |
| 240 |
245849_at |
AT5G13520
|
[peptidase M1 family protein] |
0.222 |
-0.004 |
| 241 |
264471_at |
AT1G67120
|
AtMDN1, MDN1, MIDASIN 1 |
0.222 |
0.029 |
| 242 |
258008_at |
AT3G19430
|
[late embryogenesis abundant protein-related / LEA protein-related] |
0.221 |
-0.048 |
| 243 |
263556_at |
AT2G16365
|
[F-box family protein] |
0.220 |
0.073 |
| 244 |
258894_at |
AT3G05650
|
AtRLP32, receptor like protein 32, RLP32, receptor like protein 32 |
0.220 |
0.094 |
| 245 |
260028_at |
AT1G29980
|
unknown |
0.219 |
-0.131 |
| 246 |
259173_at |
AT3G03640
|
BGLU25, beta glucosidase 25, GLUC |
0.219 |
0.040 |
| 247 |
258471_at |
AT3G06030
|
ANP3, NPK1-related protein kinase 3, AtANP3, MAPKKK12, NP3, NPK1-related protein kinase 3 |
0.218 |
-0.019 |
| 248 |
248540_at |
AT5G50160
|
ATFRO8, FERRIC REDUCTION OXIDASE 8, FRO8, ferric reduction oxidase 8 |
0.218 |
-0.031 |
| 249 |
259655_at |
AT1G55210
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.218 |
-0.120 |
| 250 |
264958_at |
AT1G76960
|
unknown |
0.218 |
-0.002 |
| 251 |
245502_at |
AT4G15640
|
unknown |
0.217 |
-0.056 |
| 252 |
260618_at |
AT1G53230
|
TCP3, TEOSINTE BRANCHED 1, cycloidea and PCF transcription factor 3 |
0.217 |
-0.155 |
| 253 |
255751_at |
AT1G31950
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
0.217 |
-0.010 |
| 254 |
253437_at |
AT4G32460
|
unknown |
0.216 |
-0.298 |
| 255 |
259731_at |
AT1G77460
|
CSI3, CELLULOSE SYNTHASE INTERACTIVE 3 |
0.216 |
-0.092 |
| 256 |
252625_at |
AT3G44750
|
ATHD2A, HD2A, HISTONE DEACETYLASE 2A, HDA3, histone deacetylase 3, HDT1 |
0.216 |
0.043 |
| 257 |
266050_at |
AT2G40770
|
[zinc ion binding] |
0.216 |
-0.000 |
| 258 |
249572_at |
AT5G37630
|
EMB2656, EMBRYO DEFECTIVE 2656 |
0.216 |
0.051 |
| 259 |
249567_at |
AT5G38020
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.216 |
0.050 |
| 260 |
260541_at |
AT2G43530
|
[Scorpion toxin-like knottin superfamily protein] |
0.216 |
0.300 |
| 261 |
266687_at |
AT2G19670
|
ATPRMT1A, ARABIDOPSIS THALIANA PROTEIN ARGININE METHYLTRANSFERASE 1A, PRMT1A, protein arginine methyltransferase 1A |
0.215 |
-0.002 |
| 262 |
261633_at |
AT1G49930
|
unknown |
0.215 |
0.024 |
| 263 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
0.215 |
-0.151 |
| 264 |
261070_at |
AT1G07390
|
AtRLP1, receptor like protein 1, RLP1, receptor like protein 1 |
0.215 |
-0.007 |
| 265 |
257696_at |
AT3G12690
|
AGC1.5, AGC kinase 1.5 |
0.214 |
0.010 |
| 266 |
261653_at |
AT1G01900
|
ATSBT1.1, SBTI1.1 |
0.214 |
-0.074 |
| 267 |
245389_at |
AT4G17480
|
[alpha/beta-Hydrolases superfamily protein] |
0.214 |
0.001 |
| 268 |
249303_at |
AT5G41460
|
unknown |
0.213 |
-0.041 |
| 269 |
246994_at |
AT5G67460
|
[O-Glycosyl hydrolases family 17 protein] |
0.213 |
-0.037 |
| 270 |
253866_at |
AT4G27480
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
0.213 |
0.009 |
| 271 |
265474_at |
AT2G15690
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.212 |
0.005 |
| 272 |
254234_at |
AT4G23680
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.212 |
-0.135 |
| 273 |
257398_at |
AT2G01990
|
unknown |
0.212 |
-0.039 |
| 274 |
250372_at |
AT5G11460
|
unknown |
0.212 |
-0.092 |
| 275 |
249019_at |
AT5G44780
|
MORF4, multiple organellar RNA editing factor 4 |
0.212 |
-0.056 |
| 276 |
266052_at |
AT2G40740
|
ATWRKY55, WRKY DNA-BINDING PROTEIN 55, WRKY55, WRKY DNA-binding protein 55 |
0.212 |
-0.083 |
| 277 |
246991_at |
AT5G67400
|
RHS19, root hair specific 19 |
0.212 |
0.055 |
| 278 |
256425_at |
AT1G33560
|
ADR1, ACTIVATED DISEASE RESISTANCE 1 |
0.212 |
0.055 |
| 279 |
260438_at |
AT1G68290
|
ENDO2, endonuclease 2 |
0.211 |
0.297 |
| 280 |
263707_at |
AT1G09300
|
[Metallopeptidase M24 family protein] |
0.211 |
-0.100 |
| 281 |
249877_at |
AT5G23150
|
HUA2, ENHANCER OF AG-4 2 |
0.211 |
-0.004 |
| 282 |
251857_at |
AT3G54770
|
ARP1, ABA-regulated RNA-binding protein 1 |
0.211 |
-0.008 |
| 283 |
247604_at |
AT5G60950
|
COBL5, COBRA-like protein 5 precursor |
0.211 |
-0.047 |
| 284 |
254174_at |
AT4G24120
|
ATYSL1, YELLOW STRIPE LIKE 1, YSL1, YELLOW STRIPE like 1 |
0.211 |
-0.022 |
| 285 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.210 |
-0.051 |
| 286 |
252716_at |
AT3G43920
|
ATDCL3, DICER-LIKE 3, DCL3, dicer-like 3 |
0.210 |
-0.030 |
| 287 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
0.210 |
-0.037 |
| 288 |
257149_at |
AT3G27280
|
ATPHB4, prohibitin 4, PHB4, prohibitin 4 |
0.210 |
-0.093 |
| 289 |
253445_at |
AT4G32605
|
[Mitochondrial glycoprotein family protein] |
0.209 |
-0.035 |
| 290 |
253681_at |
AT4G29630
|
[Cytidine/deoxycytidylate deaminase family protein] |
0.209 |
0.040 |
| 291 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.209 |
-0.558 |
| 292 |
258832_at |
AT3G07070
|
[Protein kinase superfamily protein] |
0.209 |
0.037 |
| 293 |
261500_at |
AT1G28400
|
unknown |
0.208 |
-0.124 |
| 294 |
248214_at |
AT5G53670
|
unknown |
0.208 |
0.032 |
| 295 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
0.208 |
-0.007 |
| 296 |
263496_at |
AT2G42570
|
TBL39, TRICHOME BIREFRINGENCE-LIKE 39 |
0.208 |
-0.017 |
| 297 |
262566_at |
AT1G34310
|
ARF12, auxin response factor 12 |
0.207 |
0.113 |
| 298 |
253930_at |
AT4G26740
|
AtCLO1, ATPXG1, ARABIDOPSIS THALIANA PEROXYGENASE 1, ATS1, seed gene 1, CLO1, CALEOSIN1 |
0.206 |
0.036 |
| 299 |
262953_at |
AT1G75670
|
[DNA-directed RNA polymerases] |
0.206 |
0.006 |
| 300 |
259307_at |
AT3G05230
|
[Signal peptidase subunit] |
0.206 |
-0.058 |
| 301 |
262212_at |
AT1G74890
|
ARR15, response regulator 15 |
-0.886 |
0.046 |
| 302 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
-0.773 |
0.348 |
| 303 |
259161_at |
AT3G01500
|
ATBCA1, BETA CARBONIC ANHYDRASE 1, ATSABP3, ARABIDOPSIS THALIANA SALICYLIC ACID-BINDING PROTEIN 3, CA1, carbonic anhydrase 1, SABP3, SALICYLIC ACID-BINDING PROTEIN 3 |
-0.750 |
0.657 |
| 304 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.710 |
0.099 |
| 305 |
245349_at |
AT4G16690
|
ATMES16, ARABIDOPSIS THALIANA METHYL ESTERASE 16, MES16, methyl esterase 16 |
-0.710 |
0.181 |
| 306 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
-0.706 |
0.025 |
| 307 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
-0.705 |
0.279 |
| 308 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
-0.702 |
0.144 |
| 309 |
257271_at |
AT3G28007
|
AtSWEET4, SWEET4 |
-0.685 |
0.141 |
| 310 |
266279_at |
AT2G29290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.685 |
-0.099 |
| 311 |
266078_at |
AT2G40670
|
ARR16, response regulator 16, RR16, response regulator 16 |
-0.669 |
-0.098 |
| 312 |
245047_at |
ATCG00020
|
PSBA, photosystem II reaction center protein A |
-0.667 |
-0.002 |
| 313 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
-0.659 |
0.003 |
| 314 |
258327_at |
AT3G22640
|
PAP85 |
-0.655 |
0.125 |
| 315 |
249927_at |
AT5G19220
|
ADG2, ADP GLUCOSE PYROPHOSPHORYLASE 2, APL1, ADP glucose pyrophosphorylase large subunit 1 |
-0.617 |
0.123 |
| 316 |
262421_at |
AT1G50290
|
unknown |
-0.612 |
0.013 |
| 317 |
255659_at |
AT4G00895
|
[ATPase, F1 complex, OSCP/delta subunit protein] |
-0.611 |
0.162 |
| 318 |
245025_at |
ATCG00130
|
ATPF |
-0.604 |
-0.157 |
| 319 |
258347_at |
AT3G17520
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.595 |
-0.015 |
| 320 |
262693_at |
AT1G62780
|
unknown |
-0.593 |
0.555 |
| 321 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
-0.592 |
0.461 |
| 322 |
250128_at |
AT5G16540
|
ZFN3, zinc finger nuclease 3 |
-0.588 |
0.058 |
| 323 |
259460_at |
AT1G44000
|
unknown |
-0.577 |
0.121 |
| 324 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
-0.573 |
0.450 |
| 325 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
-0.567 |
0.081 |
| 326 |
255088_at |
AT4G09350
|
CRRJ, CHLORORESPIRATORY REDUCTION J, DJC75, DNA J protein C75, NdhT, NADH dehydrogenase-like complex T |
-0.550 |
0.240 |
| 327 |
262175_at |
AT1G74880
|
NDH-O, NAD(P)H:plastoquinone dehydrogenase complex subunit O, NdhO, NADH dehydrogenase-like complex ) |
-0.548 |
0.234 |
| 328 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
-0.547 |
0.062 |
| 329 |
260367_at |
AT1G69760
|
unknown |
-0.545 |
-0.070 |
| 330 |
267247_at |
AT2G30170
|
PBCP, PHOTOSYSTEM II CORE PHOSPHATASE |
-0.542 |
0.141 |
| 331 |
246004_at |
AT5G20630
|
ATGER3, ARABIDOPSIS THALIANA GERMIN 3, GER3, germin 3, GLP3, GERMIN-LIKE PROTEIN 3, GLP3A, GLP3B |
-0.542 |
-0.023 |
| 332 |
246998_at |
AT5G67370
|
CGLD27, CONSERVED IN THE GREEN LINEAGE AND DIATOMS 27 |
-0.535 |
0.389 |
| 333 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
-0.529 |
0.410 |
| 334 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-0.525 |
0.257 |
| 335 |
264037_at |
AT2G03750
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.520 |
0.383 |
| 336 |
254505_at |
AT4G19985
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.518 |
0.061 |
| 337 |
263410_at |
AT2G04039
|
unknown |
-0.516 |
0.175 |
| 338 |
254634_at |
AT4G18650
|
[transcription factor-related] |
-0.511 |
-0.019 |
| 339 |
247899_at |
AT5G57345
|
unknown |
-0.507 |
0.265 |
| 340 |
258055_at |
AT3G16250
|
NDF4, NDH-dependent cyclic electron flow 1, PnsB3, Photosynthetic NDH subcomplex B 3 |
-0.504 |
0.716 |
| 341 |
254235_at |
AT4G23750
|
CRF2, cytokinin response factor 2, TMO3, TARGET OF MONOPTEROS 3 |
-0.501 |
0.015 |
| 342 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
-0.500 |
0.411 |
| 343 |
256697_at |
AT3G20660
|
AtOCT4, organic cation/carnitine transporter4, OCT4, organic cation/carnitine transporter4 |
-0.500 |
-0.085 |
| 344 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-0.498 |
-0.400 |
| 345 |
261422_at |
AT1G18730
|
NDF6, NDH dependent flow 6, PnsB4, Photosynthetic NDH subcomplex B 4 |
-0.495 |
0.458 |
| 346 |
255046_at |
AT4G09650
|
ATPD, ATP synthase delta-subunit gene, PDE332, PIGMENT DEFECTIVE 332 |
-0.491 |
0.249 |
| 347 |
245011_at |
ATCG00430
|
PSBG, photosystem II reaction center protein G |
-0.491 |
-0.177 |
| 348 |
256603_at |
AT3G28270
|
unknown |
-0.490 |
0.601 |
| 349 |
250531_at |
AT5G08650
|
[Small GTP-binding protein] |
-0.488 |
0.090 |
| 350 |
257909_at |
AT3G25480
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.486 |
0.068 |
| 351 |
256894_at |
AT3G21870
|
CYCP2;1, cyclin p2;1 |
-0.485 |
0.196 |
| 352 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
-0.471 |
-0.351 |
| 353 |
262288_at |
AT1G70760
|
CRR23, CHLORORESPIRATORY REDUCTION 23, NdhL, NADH dehydrogenase-like complex L |
-0.470 |
0.231 |
| 354 |
266778_at |
AT2G29090
|
CYP707A2, cytochrome P450, family 707, subfamily A, polypeptide 2 |
-0.467 |
0.192 |
| 355 |
266372_at |
AT2G41310
|
ARR8, RESPONSE REGULATOR 8, ATRR3, response regulator 3, RR3, response regulator 3 |
-0.464 |
0.039 |
| 356 |
245460_at |
AT4G16990
|
RLM3, RESISTANCE TO LEPTOSPHAERIA MACULANS 3 |
-0.460 |
-0.084 |
| 357 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
-0.460 |
0.210 |
| 358 |
254145_at |
AT4G24700
|
unknown |
-0.459 |
0.104 |
| 359 |
262526_at |
AT1G17050
|
AtSPS2, SPS2, solanesyl diphosphate synthase 2 |
-0.458 |
0.258 |
| 360 |
262945_at |
AT1G79510
|
[Uncharacterized conserved protein (DUF2358)] |
-0.458 |
0.199 |
| 361 |
254175_at |
AT4G24050
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.457 |
-0.030 |
| 362 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
-0.455 |
0.210 |
| 363 |
252441_at |
AT3G46780
|
PTAC16, plastid transcriptionally active 16 |
-0.454 |
0.353 |
| 364 |
249454_at |
AT5G39520
|
unknown |
-0.452 |
0.093 |
| 365 |
252463_at |
AT3G47070
|
[LOCATED IN: thylakoid, chloroplast thylakoid membrane, chloroplast, chloroplast envelope] |
-0.446 |
0.173 |
| 366 |
259275_at |
AT3G01060
|
unknown |
-0.444 |
0.116 |
| 367 |
245233_at |
AT4G25580
|
[CAP160 protein] |
-0.444 |
0.052 |
| 368 |
253849_at |
AT4G28080
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.444 |
0.432 |
| 369 |
256427_at |
AT3G11090
|
LBD21, LOB domain-containing protein 21 |
-0.441 |
0.093 |
| 370 |
266600_at |
AT2G46070
|
ATMPK12, MAPK12, MPK12, mitogen-activated protein kinase 12 |
-0.440 |
0.025 |
| 371 |
248537_at |
AT5G50100
|
[Putative thiol-disulphide oxidoreductase DCC] |
-0.435 |
0.159 |
| 372 |
260744_at |
AT1G15010
|
unknown |
-0.435 |
0.067 |
| 373 |
258227_at |
AT3G15620
|
UVR3, UV REPAIR DEFECTIVE 3 |
-0.433 |
-0.130 |
| 374 |
247455_at |
AT5G62470
|
ATMYB96, MYB DOMAIN PROTEIN 96, MYB96, myb domain protein 96, MYBCOV1 |
-0.430 |
-0.068 |
| 375 |
250960_at |
AT5G02940
|
unknown |
-0.430 |
0.294 |
| 376 |
259677_at |
AT1G77740
|
PIP5K2, phosphatidylinositol-4-phosphate 5-kinase 2 |
-0.429 |
0.042 |
| 377 |
263653_at |
AT1G04310
|
ERS2, ethylene response sensor 2 |
-0.426 |
-0.116 |
| 378 |
263142_at |
AT1G65230
|
[Uncharacterized conserved protein (DUF2358)] |
-0.426 |
0.291 |
| 379 |
261788_at |
AT1G15980
|
NDF1, NDH-dependent cyclic electron flow 1, NDH48, NAD(P)H DEHYDROGENASE SUBUNIT 48, PnsB1, Photosynthetic NDH subcomplex B 1 |
-0.423 |
0.251 |
| 380 |
250006_at |
AT5G18660
|
PCB2, PALE-GREEN AND CHLOROPHYLL B REDUCED 2 |
-0.422 |
0.252 |
| 381 |
254384_at |
AT4G21870
|
[HSP20-like chaperones superfamily protein] |
-0.422 |
-0.097 |
| 382 |
263067_at |
AT2G17550
|
TRM26, TON1 Recruiting Motif 26 |
-0.421 |
0.048 |
| 383 |
249694_at |
AT5G35790
|
G6PD1, glucose-6-phosphate dehydrogenase 1 |
-0.421 |
0.290 |
| 384 |
259981_at |
AT1G76450
|
[Photosystem II reaction center PsbP family protein] |
-0.421 |
0.196 |
| 385 |
266544_at |
AT2G35300
|
AtLEA4-2, late embryogenesis abundant 4-2, LEA18, late embryogenesis abundant 18, LEA4-2, late embryogenesis abundant 4-2 |
-0.420 |
0.197 |
| 386 |
259707_at |
AT1G77490
|
TAPX, thylakoidal ascorbate peroxidase |
-0.418 |
0.253 |
| 387 |
249932_at |
AT5G22390
|
unknown |
-0.416 |
-0.058 |
| 388 |
253009_at |
AT4G37930
|
SHM1, serine transhydroxymethyltransferase 1, SHMT1, SERINE HYDROXYMETHYLTRANSFERASE 1, STM, SERINE TRANSHYDROXYMETHYLTRANSFERASE |
-0.411 |
0.137 |
| 389 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
-0.409 |
0.030 |
| 390 |
249117_at |
AT5G43840
|
AT-HSFA6A, heat shock transcription factor A6A, HSFA6A, heat shock transcription factor A6A |
-0.407 |
0.029 |
| 391 |
261769_at |
AT1G76100
|
PETE1, plastocyanin 1 |
-0.407 |
0.941 |
| 392 |
266979_at |
AT2G39470
|
PnsL1, Photosynthetic NDH subcomplex L 1, PPL2, PsbP-like protein 2 |
-0.405 |
0.320 |
| 393 |
245007_at |
ATCG00350
|
PSAA |
-0.403 |
-0.225 |
| 394 |
259129_at |
AT3G02150
|
PTF1, plastid transcription factor 1, TCP13, TEOSINTE BRANCHED1, CYCLOIDEA AND PCF TRANSCRIPTION FACTOR 13, TFPD |
-0.403 |
0.078 |
| 395 |
250739_at |
AT5G05740
|
ATEGY2, EGY2, ethylene-dependent gravitropism-deficient and yellow-green-like 2 |
-0.403 |
0.138 |
| 396 |
250257_at |
AT5G13770
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
-0.403 |
0.195 |
| 397 |
259970_at |
AT1G76570
|
AtLHCB7, LHCB7, light-harvesting complex B7 |
-0.401 |
0.217 |
| 398 |
261247_at |
AT1G20070
|
unknown |
-0.400 |
0.158 |
| 399 |
264442_at |
AT1G27480
|
[alpha/beta-Hydrolases superfamily protein] |
-0.395 |
0.266 |
| 400 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
-0.394 |
-0.023 |
| 401 |
266815_at |
AT2G44900
|
ARABIDILLO-1, ARABIDILLO-1, ARABIDILLO1, F-box Armadillo protein 1 |
-0.394 |
0.151 |
| 402 |
259616_at |
AT1G47960
|
ATC/VIF1, CELL WALL / VACUOLAR INHIBITOR OF FRUCTOSIDASE 1, C/VIF1, cell wall / vacuolar inhibitor of fructosidase 1 |
-0.393 |
0.257 |
| 403 |
263593_at |
AT2G01860
|
EMB975, EMBRYO DEFECTIVE 975 |
-0.392 |
0.078 |
| 404 |
252092_at |
AT3G51420
|
ATSSL4, STRICTOSIDINE SYNTHASE-LIKE 4, SSL4, strictosidine synthase-like 4 |
-0.391 |
0.118 |
| 405 |
250262_at |
AT5G13410
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.390 |
0.344 |
| 406 |
255719_at |
AT1G32080
|
AtLrgB, LrgB |
-0.389 |
0.133 |
| 407 |
253823_at |
AT4G28030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.389 |
0.227 |
| 408 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
-0.389 |
0.100 |
| 409 |
263495_at |
AT2G42530
|
COR15B, cold regulated 15b |
-0.389 |
0.554 |
| 410 |
251157_at |
AT3G63140
|
CSP41A, chloroplast stem-loop binding protein of 41 kDa |
-0.387 |
0.414 |
| 411 |
250165_at |
AT5G15290
|
CASP5, Casparian strip membrane domain protein 5 |
-0.387 |
-0.080 |
| 412 |
257294_at |
AT3G15570
|
[Phototropic-responsive NPH3 family protein] |
-0.386 |
-0.144 |
| 413 |
262768_at |
AT1G12990
|
[beta-1,4-N-acetylglucosaminyltransferase family protein] |
-0.384 |
-0.016 |
| 414 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
-0.384 |
0.166 |
| 415 |
254638_at |
AT4G18740
|
[Rho termination factor] |
-0.382 |
0.336 |
| 416 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
-0.382 |
0.100 |
| 417 |
245776_at |
AT1G30260
|
unknown |
-0.380 |
0.093 |
| 418 |
266572_at |
AT2G23840
|
[HNH endonuclease] |
-0.380 |
0.091 |
| 419 |
257791_at |
AT3G27110
|
[Peptidase family M48 family protein] |
-0.379 |
0.112 |
| 420 |
265084_at |
AT1G03790
|
AtTZF4, SOM, SOMNUS, TZF4, Tandem CCCH Zinc Finger protein 4 |
-0.377 |
0.079 |
| 421 |
262412_at |
AT1G34760
|
GF14 OMICRON, GRF11, general regulatory factor 11, RHS5, ROOT HAIR SPECIFIC 5 |
-0.377 |
0.056 |
| 422 |
253956_at |
AT4G26700
|
ATFIM1, ARABIDOPSIS THALIANA FIMBRIN 1, FIM1, fimbrin 1 |
-0.377 |
0.180 |
| 423 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
-0.375 |
0.038 |
| 424 |
259432_at |
AT1G01520
|
ASG4, ALTERED SEED GERMINATION 4 |
-0.373 |
0.273 |
| 425 |
256611_at |
AT3G29270
|
[RING/U-box superfamily protein] |
-0.372 |
0.459 |
| 426 |
250884_at |
AT5G03940
|
54CP, 54 CHLOROPLAST PROTEIN, CPSRP54, chloroplast signal recognition particle 54 kDa subunit, FFC, FIFTY-FOUR CHLOROPLAST HOMOLOGUE, SRP54CP, SIGNAL RECOGNITION PARTICLE 54 KDA SUBUNIT CHLOROPLAST PROTEIN |
-0.372 |
0.268 |
| 427 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
-0.371 |
-0.025 |
| 428 |
249035_at |
AT5G44190
|
ATGLK2, GLK2, GOLDEN2-like 2, GPRI2, GBF'S PRO-RICH REGION-INTERACTING FACTOR 2 |
-0.369 |
0.505 |
| 429 |
264597_at |
AT1G04620
|
HCAR, 7-hydroxymethyl chlorophyll a (HMChl) reductase |
-0.369 |
0.086 |
| 430 |
264201_at |
AT1G22630
|
unknown |
-0.368 |
0.290 |
| 431 |
245242_at |
AT1G44446
|
ATCAO, ARABIDOPSIS THALIANA CHLOROPHYLL A OXYGENASE, CAO, CHLOROPHYLL A OXYGENASE, CH1, CHLORINA 1 |
-0.365 |
0.253 |
| 432 |
266278_at |
AT2G29300
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.364 |
-0.027 |
| 433 |
250255_at |
AT5G13730
|
SIG4, sigma factor 4, SIGD |
-0.364 |
0.400 |
| 434 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
-0.364 |
0.141 |
| 435 |
266329_at |
AT2G01590
|
CRR3, CHLORORESPIRATORY REDUCTION 3 |
-0.363 |
0.272 |
| 436 |
253517_at |
AT4G31390
|
ABC1K1, ABC1-like kinase 1, ACDO1, ABC1-like kinase related to chlorophyll degradation and oxidative stress 1, AtACDO1, PGR6, PROTON GRADIENT REGULATION 6 |
-0.363 |
0.161 |
| 437 |
257772_at |
AT3G23080
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.361 |
0.210 |
| 438 |
263676_at |
AT1G09340
|
CRB, chloroplast RNA binding, CSP41B, CHLOROPLAST STEM-LOOP BINDING PROTEIN OF 41 KDA, HIP1.3, heteroglycan-interacting protein 1.3 |
-0.360 |
0.262 |
| 439 |
245592_at |
AT4G14540
|
NF-YB3, nuclear factor Y, subunit B3 |
-0.356 |
0.174 |
| 440 |
267505_at |
AT2G45560
|
CYP76C1, cytochrome P450, family 76, subfamily C, polypeptide 1 |
-0.354 |
0.117 |
| 441 |
249047_at |
AT5G44410
|
[FAD-binding Berberine family protein] |
-0.353 |
0.022 |
| 442 |
254398_at |
AT4G21280
|
PSBQ, PHOTOSYSTEM II SUBUNIT Q, PSBQ-1, PHOTOSYSTEM II SUBUNIT Q-1, PSBQA, photosystem II subunit QA |
-0.351 |
0.244 |
| 443 |
245002_at |
ATCG00270
|
PSBD, photosystem II reaction center protein D |
-0.351 |
-0.119 |
| 444 |
262313_at |
AT1G70900
|
unknown |
-0.351 |
0.453 |
| 445 |
253381_at |
AT4G33350
|
AtTic22-IV, translocon at the inner envelope membrane of chloroplasts 22-IV, Tic22-IV, translocon at the inner envelope membrane of chloroplasts 22-IV |
-0.351 |
0.133 |
| 446 |
265939_at |
AT2G19650
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.350 |
0.078 |
| 447 |
246313_at |
AT1G31920
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.348 |
0.191 |
| 448 |
249510_at |
AT5G38510
|
[Rhomboid-related intramembrane serine protease family protein] |
-0.348 |
0.224 |
| 449 |
253048_at |
AT4G37560
|
[Acetamidase/Formamidase family protein] |
-0.347 |
0.478 |
| 450 |
244932_at |
ATCG01060
|
PSAC |
-0.346 |
-0.205 |
| 451 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
-0.344 |
0.063 |
| 452 |
255440_at |
AT4G02530
|
[chloroplast thylakoid lumen protein] |
-0.341 |
0.176 |
| 453 |
247891_at |
AT5G57960
|
[GTP-binding protein, HflX] |
-0.341 |
0.134 |
| 454 |
251762_at |
AT3G55800
|
SBPASE, sedoheptulose-bisphosphatase |
-0.340 |
0.244 |
| 455 |
250420_at |
AT5G11260
|
HY5, ELONGATED HYPOCOTYL 5, TED 5, REVERSAL OF THE DET PHENOTYPE 5 |
-0.339 |
0.074 |
| 456 |
250624_at |
AT5G07330
|
unknown |
-0.339 |
-0.057 |
| 457 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
-0.338 |
0.077 |
| 458 |
251647_at |
AT3G57770
|
[Protein kinase superfamily protein] |
-0.338 |
0.096 |
| 459 |
247793_at |
AT5G58650
|
PSY1, plant peptide containing sulfated tyrosine 1 |
-0.338 |
0.038 |
| 460 |
251916_at |
AT3G53960
|
[Major facilitator superfamily protein] |
-0.337 |
0.070 |
| 461 |
260266_at |
AT1G68520
|
BBX14, B-box domain protein 14 |
-0.337 |
0.202 |
| 462 |
250327_at |
AT5G12050
|
unknown |
-0.337 |
-0.219 |
| 463 |
246226_at |
AT4G37200
|
HCF164, HIGH CHLOROPHYLL FLUORESCENCE 164 |
-0.336 |
0.130 |
| 464 |
263127_at |
AT1G78610
|
MSL6, mechanosensitive channel of small conductance-like 6 |
-0.333 |
-0.133 |
| 465 |
257827_at |
AT3G26630
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.332 |
0.141 |
| 466 |
248293_at |
AT5G53050
|
[alpha/beta-Hydrolases superfamily protein] |
-0.331 |
0.303 |
| 467 |
265628_at |
AT2G27290
|
unknown |
-0.331 |
0.195 |
| 468 |
259927_at |
AT1G75100
|
JAC1, J-domain protein required for chloroplast accumulation response 1 |
-0.331 |
0.054 |
| 469 |
258819_at |
AT3G04590
|
AHL14, AT-hook motif nuclear localized protein 14 |
-0.331 |
-0.041 |
| 470 |
258375_at |
AT3G17470
|
ATCRSH, CRSH, Ca2+-activated RelA/spot homolog |
-0.330 |
0.128 |
| 471 |
255637_at |
AT4G00750
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.330 |
0.184 |
| 472 |
255285_at |
AT4G04630
|
unknown |
-0.329 |
0.023 |
| 473 |
248969_at |
AT5G45310
|
unknown |
-0.328 |
0.027 |
| 474 |
255905_at |
AT1G17810
|
BETA-TIP, beta-tonoplast intrinsic protein |
-0.327 |
-0.029 |
| 475 |
260542_at |
AT2G43560
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.327 |
0.199 |
| 476 |
245213_at |
AT1G44575
|
CP22, NPQ4, NONPHOTOCHEMICAL QUENCHING 4, PSBS, PHOTOSYSTEM II SUBUNIT S |
-0.327 |
0.389 |
| 477 |
251155_at |
AT3G63160
|
OEP6, outer envelope protein 6 |
-0.325 |
0.233 |
| 478 |
258049_at |
AT3G16220
|
[Predicted eukaryotic LigT] |
-0.324 |
0.126 |
| 479 |
262940_at |
AT1G79520
|
[Cation efflux family protein] |
-0.324 |
0.278 |
| 480 |
257999_at |
AT3G27540
|
[beta-1,4-N-acetylglucosaminyltransferase family protein] |
-0.323 |
0.035 |
| 481 |
245346_at |
AT4G17090
|
AtBAM3, BAM3, BETA-AMYLASE 3, BMY8, BETA-AMYLASE 8, CT-BMY, chloroplast beta-amylase |
-0.322 |
0.230 |
| 482 |
266813_at |
AT2G44920
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.321 |
0.136 |
| 483 |
245165_at |
AT2G33180
|
unknown |
-0.321 |
0.112 |
| 484 |
253790_at |
AT4G28660
|
PSB28, photosystem II reaction center PSB28 protein |
-0.320 |
0.151 |
| 485 |
250555_at |
AT5G07950
|
unknown |
-0.319 |
0.104 |
| 486 |
248402_at |
AT5G52100
|
CRR1, chlororespiration reduction 1 |
-0.319 |
0.126 |
| 487 |
252100_at |
AT3G51110
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.319 |
-0.090 |
| 488 |
264700_at |
AT1G70100
|
unknown |
-0.319 |
0.010 |
| 489 |
264096_at |
AT1G78995
|
unknown |
-0.318 |
0.154 |
| 490 |
261112_at |
AT1G75420
|
[UDP-Glycosyltransferase superfamily protein] |
-0.318 |
-0.031 |
| 491 |
262916_at |
AT1G59700
|
ATGSTU16, glutathione S-transferase TAU 16, GSTU16, glutathione S-transferase TAU 16 |
-0.318 |
0.105 |
| 492 |
263946_at |
AT2G36000
|
EMB3114, EMBRYO DEFECTIVE 3114 |
-0.317 |
0.012 |
| 493 |
259791_at |
AT1G29700
|
[Metallo-hydrolase/oxidoreductase superfamily protein] |
-0.315 |
0.347 |
| 494 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
-0.315 |
0.045 |
| 495 |
261118_at |
AT1G75460
|
[ATP-dependent protease La (LON) domain protein] |
-0.315 |
0.390 |
| 496 |
267010_at |
AT2G39250
|
SNZ, SCHNARCHZAPFEN |
-0.315 |
-0.061 |
| 497 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
-0.314 |
0.054 |
| 498 |
249876_at |
AT5G23060
|
CaS, calcium sensing receptor |
-0.314 |
0.338 |
| 499 |
246483_at |
AT5G16000
|
AtNIK1, NIK1, NSP-interacting kinase 1 |
-0.313 |
-0.029 |
| 500 |
251699_at |
AT3G56630
|
CYP94D2, cytochrome P450, family 94, subfamily D, polypeptide 2 |
-0.312 |
0.058 |
| 501 |
267298_at |
AT2G23760
|
BLH4, BEL1-like homeodomain 4, SAW2, SAWTOOTH 2 |
-0.312 |
-0.058 |
| 502 |
253597_at |
AT4G30690
|
[Translation initiation factor 3 protein] |
-0.312 |
0.124 |
| 503 |
265249_at |
AT2G01940
|
ATIDD15, ARABIDOPSIS THALIANA INDETERMINATE(ID)-DOMAIN 15, SGR5, SHOOT GRAVITROPISM 5 |
-0.312 |
-0.008 |
| 504 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.312 |
0.089 |
| 505 |
253197_at |
AT4G35250
|
HCF244, high chlorophyll fluorescence phenotype 244 |
-0.311 |
0.164 |
| 506 |
261026_at |
AT1G01240
|
unknown |
-0.311 |
0.194 |
| 507 |
255720_at |
AT1G32060
|
PRK, phosphoribulokinase |
-0.310 |
0.249 |
| 508 |
260989_at |
AT1G53450
|
unknown |
-0.310 |
0.029 |
| 509 |
260205_at |
AT1G70700
|
JAZ9, JASMONATE-ZIM-DOMAIN PROTEIN 9, TIFY7 |
-0.310 |
0.122 |
| 510 |
265840_at |
AT2G14530
|
TBL13, TRICHOME BIREFRINGENCE-LIKE 13 |
-0.310 |
-0.042 |
| 511 |
250095_at |
AT5G17230
|
PSY, PHYTOENE SYNTHASE |
-0.309 |
0.258 |
| 512 |
266277_at |
AT2G29310
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.308 |
0.159 |
| 513 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.308 |
0.155 |
| 514 |
245701_at |
AT5G04140
|
FD-GOGAT, FERREDOXIN-DEPENDENT GLUTAMATE SYNTHASE, GLS1, FERREDOXIN-DEPENDENT GLUTAMATE SYNTHASE 1, GLU1, glutamate synthase 1, GLUS |
-0.307 |
0.091 |
| 515 |
258901_at |
AT3G05640
|
[Protein phosphatase 2C family protein] |
-0.307 |
0.187 |
| 516 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
-0.307 |
-0.182 |
| 517 |
258333_at |
AT3G16000
|
MFP1, MAR binding filament-like protein 1 |
-0.307 |
0.303 |
| 518 |
253224_at |
AT4G34860
|
A/N-InvB, alkaline/neutral invertase B |
-0.305 |
0.136 |
| 519 |
252290_at |
AT3G49140
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.304 |
0.106 |
| 520 |
254553_at |
AT4G19530
|
[disease resistance protein (TIR-NBS-LRR class) family] |
-0.304 |
0.213 |
| 521 |
250563_at |
AT5G08050
|
unknown |
-0.303 |
0.150 |
| 522 |
259466_at |
AT1G19050
|
ARR7, response regulator 7 |
-0.303 |
-0.051 |
| 523 |
258181_at |
AT3G21670
|
AtNPF6.4, NPF6.4, NRT1/ PTR family 6.4 |
-0.302 |
0.119 |
| 524 |
262604_at |
AT1G15060
|
[Uncharacterised conserved protein UCP031088, alpha/beta hydrolase] |
-0.302 |
0.026 |
| 525 |
260877_at |
AT1G21500
|
unknown |
-0.301 |
0.678 |
| 526 |
267080_at |
AT2G41190
|
[Transmembrane amino acid transporter family protein] |
-0.301 |
0.111 |
| 527 |
251856_at |
AT3G54720
|
AMP1, ALTERED MERISTEM PROGRAM 1, AtAMP1, COP2, CONSTITUTIVE MORPHOGENESIS 2, HPT, HAUPTLING, MFO1, Multifolia, PT, PRIMORDIA TIMING |
-0.301 |
0.009 |
| 528 |
262309_at |
AT1G70820
|
[phosphoglucomutase, putative / glucose phosphomutase, putative] |
-0.300 |
0.289 |
| 529 |
245015_at |
ATCG00490
|
RBCL |
-0.300 |
-0.126 |
| 530 |
247714_at |
AT5G59340
|
WOX2, WUSCHEL related homeobox 2 |
-0.299 |
0.068 |
| 531 |
261205_at |
AT1G12790
|
unknown |
-0.298 |
0.077 |
| 532 |
266690_at |
AT2G19900
|
ATNADP-ME1, Arabidopsis thaliana NADP-malic enzyme 1, NADP-ME1, NADP-malic enzyme 1 |
-0.298 |
0.059 |
| 533 |
262952_at |
AT1G75770
|
unknown |
-0.298 |
0.192 |
| 534 |
258080_at |
AT3G25930
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.298 |
-0.052 |
| 535 |
250340_at |
AT5G11840
|
unknown |
-0.297 |
0.124 |
| 536 |
263668_at |
AT1G04350
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.297 |
0.034 |
| 537 |
251066_at |
AT5G01880
|
DAFL2, DAF-Like gene 2 |
-0.297 |
0.066 |
| 538 |
264959_at |
AT1G77090
|
[Mog1/PsbP/DUF1795-like photosystem II reaction center PsbP family protein] |
-0.297 |
0.181 |
| 539 |
260704_at |
AT1G32470
|
[Single hybrid motif superfamily protein] |
-0.297 |
0.229 |
| 540 |
262199_at |
AT1G53800
|
unknown |
-0.296 |
-0.042 |
| 541 |
266322_at |
AT2G46690
|
SAUR32, SMALL AUXIN UPREGULATED RNA 32 |
-0.296 |
-0.071 |
| 542 |
251218_at |
AT3G62410
|
CP12, CP12 DOMAIN-CONTAINING PROTEIN 1, CP12-2, CP12 domain-containing protein 2 |
-0.295 |
0.413 |
| 543 |
264799_at |
AT1G08550
|
AVDE1, ARABIDOPSIS VIOLAXANTHIN DE-EPOXIDASE 1, NPQ1, non-photochemical quenching 1 |
-0.295 |
0.152 |
| 544 |
262373_at |
AT1G73120
|
unknown |
-0.295 |
-0.133 |
| 545 |
261782_at |
AT1G76110
|
[HMG (high mobility group) box protein with ARID/BRIGHT DNA-binding domain] |
-0.295 |
0.304 |
| 546 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
-0.294 |
-0.235 |
| 547 |
255878_at |
AT2G40620
|
[Basic-leucine zipper (bZIP) transcription factor family protein] |
-0.293 |
-0.012 |
| 548 |
266767_at |
AT2G46910
|
[Plastid-lipid associated protein PAP / fibrillin family protein] |
-0.292 |
0.090 |
| 549 |
250161_at |
AT5G15240
|
[Transmembrane amino acid transporter family protein] |
-0.292 |
0.006 |
| 550 |
246062_at |
AT5G19330
|
ARIA, ARM repeat protein interacting with ABF2 |
-0.292 |
0.192 |
| 551 |
260567_at |
AT2G43820
|
ATSAGT1, Arabidopsis thaliana salicylic acid glucosyltransferase 1, GT, SAGT1, salicylic acid glucosyltransferase 1, SGT1, UDP-glucose:salicylic acid glucosyltransferase 1, UGT74F2, UDP-glucosyltransferase 74F2 |
-0.290 |
0.032 |
| 552 |
245936_at |
AT5G19850
|
[alpha/beta-Hydrolases superfamily protein] |
-0.290 |
0.165 |
| 553 |
264435_at |
AT1G10360
|
ATGSTU18, glutathione S-transferase TAU 18, GST29, GLUTATHIONE S-TRANSFERASE 29, GSTU18, glutathione S-transferase TAU 18 |
-0.289 |
0.292 |
| 554 |
246298_at |
AT3G51770
|
ATEOL1, ARABIDOPSIS ETHYLENE OVERPRODUCER 1, ETO1, ETHYLENE OVERPRODUCER 1 |
-0.289 |
-0.106 |
| 555 |
258456_at |
AT3G22420
|
ATWNK2, ARABIDOPSIS THALIANA WITH NO K 2, WNK2, with no lysine (K) kinase 2, ZIK3 |
-0.289 |
0.180 |
| 556 |
251181_at |
AT3G62820
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.289 |
-0.039 |
| 557 |
244975_at |
ATCG00710
|
PSBH, photosystem II reaction center protein H |
-0.289 |
-0.117 |
| 558 |
260784_at |
AT1G06180
|
ATMYB13, myb domain protein 13, ATMYBLFGN, MYB13, myb domain protein 13 |
-0.288 |
0.137 |
| 559 |
255579_at |
AT4G01460
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.287 |
-0.084 |
| 560 |
250158_at |
AT5G15190
|
unknown |
-0.287 |
0.022 |
| 561 |
255866_at |
AT2G30350
|
[Excinuclease ABC, C subunit, N-terminal] |
-0.287 |
0.100 |
| 562 |
245003_at |
ATCG00280
|
PSBC, photosystem II reaction center protein C |
-0.287 |
-0.062 |
| 563 |
253387_at |
AT4G33010
|
AtGLDP1, glycine decarboxylase P-protein 1, GLDP1, glycine decarboxylase P-protein 1 |
-0.286 |
0.148 |
| 564 |
255328_at |
AT4G04350
|
EMB2369, EMBRYO DEFECTIVE 2369 |
-0.286 |
0.092 |
| 565 |
246231_at |
AT4G37080
|
unknown |
-0.286 |
-0.059 |
| 566 |
246125_at |
AT5G19875
|
unknown |
-0.286 |
-0.018 |
| 567 |
264238_at |
AT1G54740
|
unknown |
-0.286 |
0.029 |
| 568 |
256514_at |
AT1G66130
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.286 |
0.178 |
| 569 |
262612_at |
AT1G14150
|
PnsL2, Photosynthetic NDH subcomplex L 2, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.285 |
0.506 |
| 570 |
261952_at |
AT1G64430
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.285 |
0.097 |
| 571 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
-0.285 |
-0.094 |
| 572 |
267535_at |
AT2G41940
|
ZFP8, zinc finger protein 8 |
-0.285 |
-0.015 |
| 573 |
259234_at |
AT3G11620
|
[alpha/beta-Hydrolases superfamily protein] |
-0.285 |
0.062 |
| 574 |
253741_at |
AT4G28890
|
[RING/U-box superfamily protein] |
-0.284 |
-0.031 |
| 575 |
261218_at |
AT1G20020
|
ATLFNR2, LEAF FNR 2, FNR2, ferredoxin-NADP(+)-oxidoreductase 2, LFNR2, leaf-type chloroplast-targeted FNR 2 |
-0.283 |
0.225 |
| 576 |
249120_at |
AT5G43750
|
NDH18, NAD(P)H dehydrogenase 18, PnsB5, Photosynthetic NDH subcomplex B 5 |
-0.283 |
0.318 |
| 577 |
264124_at |
AT1G79360
|
ATOCT2, organic cation/carnitine transporter 2, OCT2, ORGANIC CATION TRANSPORTER 2, OCT2, organic cation/carnitine transporter 2 |
-0.282 |
-0.072 |
| 578 |
255826_at |
AT2G40490
|
HEME2 |
-0.282 |
0.125 |
| 579 |
245352_at |
AT4G15490
|
UGT84A3 |
-0.282 |
0.154 |
| 580 |
248224_at |
AT5G53490
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.282 |
0.205 |
| 581 |
255787_at |
AT2G33590
|
AtCRL1, CRL1, CCR(Cinnamoyl coA:NADP oxidoreductase)-like 1 |
-0.282 |
0.055 |
| 582 |
259179_at |
AT3G01660
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.282 |
0.184 |
| 583 |
247024_at |
AT5G66985
|
unknown |
-0.280 |
0.355 |
| 584 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
-0.280 |
-0.030 |
| 585 |
263533_at |
AT2G24820
|
AtTic55, translocon at the inner envelope membrane of chloroplasts 55, Tic55, translocon at the inner envelope membrane of chloroplasts 55, TIC55-II, translocon at the inner envelope membrane of chloroplasts 55-II |
-0.279 |
0.103 |
| 586 |
248962_at |
AT5G45680
|
ATFKBP13, FK506 BINDING PROTEIN 13, FKBP13, FK506-binding protein 13 |
-0.279 |
0.145 |
| 587 |
267294_at |
AT2G23670
|
YCF37, homolog of Synechocystis YCF37 |
-0.279 |
0.232 |
| 588 |
261208_at |
AT1G12930
|
[ARM repeat superfamily protein] |
-0.279 |
-0.023 |
| 589 |
260106_at |
AT1G35420
|
[alpha/beta-Hydrolases superfamily protein] |
-0.278 |
0.143 |
| 590 |
262626_at |
AT1G06430
|
FTSH8, FTSH protease 8 |
-0.278 |
0.148 |
| 591 |
252081_at |
AT3G51910
|
AT-HSFA7A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A7A, HSFA7A, heat shock transcription factor A7A |
-0.278 |
-0.055 |
| 592 |
253530_at |
AT4G31530
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.278 |
0.211 |
| 593 |
261519_at |
AT1G71810
|
[Protein kinase superfamily protein] |
-0.277 |
0.164 |
| 594 |
258025_at |
AT3G19480
|
3-PGDH, PGDH3, phosphoglycerate dehydrogenase 3 |
-0.277 |
0.604 |
| 595 |
257932_at |
AT3G17040
|
HCF107, high chlorophyll fluorescent 107 |
-0.277 |
0.227 |
| 596 |
251465_at |
AT3G59360
|
ATUTR6, UDP-GALACTOSE TRANSPORTER 6, UTR6, UDP-galactose transporter 6 |
-0.276 |
-0.067 |
| 597 |
247818_at |
AT5G58370
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.276 |
0.047 |
| 598 |
261316_at |
AT1G53165
|
ATMAP4K ALPHA1 |
-0.276 |
0.004 |
| 599 |
260837_at |
AT1G43670
|
AtcFBP, Arabidopsis thaliana cytosolic fructose-1,6-bisphosphatase, FBP, fructose-1,6-bisphosphatase, FINS1, FRUCTOSE INSENSITIVE 1 |
-0.276 |
0.191 |
| 600 |
254038_at |
AT4G25910
|
ATCNFU3, NFU3, NFU domain protein 3 |
-0.274 |
0.137 |