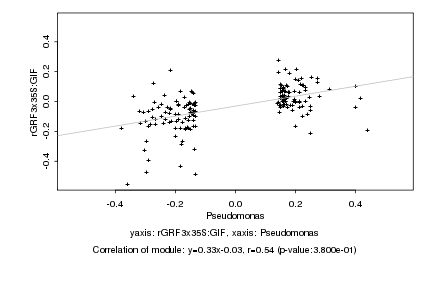
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Pseudomonas |
Log2 signal ratio
rGRF3x35S:GIF |
| 1 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
0.439 |
-0.193 |
| 2 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
0.415 |
0.022 |
| 3 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
0.400 |
-0.038 |
| 4 |
251494_at |
AT3G59350
|
[Protein kinase superfamily protein] |
0.398 |
0.103 |
| 5 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.311 |
0.082 |
| 6 |
263118_at |
AT1G03090
|
MCCA |
0.279 |
0.038 |
| 7 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
0.272 |
0.157 |
| 8 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
0.271 |
0.128 |
| 9 |
264467_at |
AT1G10140
|
[Uncharacterised conserved protein UCP031279] |
0.253 |
0.162 |
| 10 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.249 |
-0.056 |
| 11 |
253827_at |
AT4G28085
|
unknown |
0.249 |
-0.028 |
| 12 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
0.247 |
-0.212 |
| 13 |
267532_at |
AT2G42040
|
unknown |
0.246 |
0.032 |
| 14 |
261375_at |
AT1G53160
|
FTM6, FLORAL TRANSITION AT THE MERISTEM6, SPL4, squamosa promoter binding protein-like 4 |
0.240 |
-0.086 |
| 15 |
252570_at |
AT3G45300
|
ATIVD, IVD, isovaleryl-CoA-dehydrogenase, IVDH, ISOVALERYL-COA-DEHYDROGENASE |
0.233 |
0.002 |
| 16 |
254665_at |
AT4G18340
|
[Glycosyl hydrolase superfamily protein] |
0.233 |
0.096 |
| 17 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
0.231 |
0.076 |
| 18 |
250580_at |
AT5G07440
|
GDH2, glutamate dehydrogenase 2 |
0.225 |
0.110 |
| 19 |
251642_at |
AT3G57520
|
AtSIP2, seed imbibition 2, RS2, raffinose synthase 2, SIP2, seed imbibition 2 |
0.222 |
-0.095 |
| 20 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
0.221 |
0.113 |
| 21 |
259694_at |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
0.221 |
-0.029 |
| 22 |
260662_at |
AT1G19540
|
[NmrA-like negative transcriptional regulator family protein] |
0.218 |
0.155 |
| 23 |
259982_at |
AT1G76410
|
ATL8 |
0.215 |
0.114 |
| 24 |
246034_at |
AT5G08350
|
[GRAM domain-containing protein / ABA-responsive protein-related] |
0.213 |
-0.001 |
| 25 |
245076_at |
AT2G23170
|
GH3.3 |
0.212 |
0.064 |
| 26 |
256965_at |
AT3G13450
|
DIN4, DARK INDUCIBLE 4 |
0.212 |
0.001 |
| 27 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
0.211 |
-0.036 |
| 28 |
265536_at |
AT2G15880
|
[Leucine-rich repeat (LRR) family protein] |
0.207 |
0.141 |
| 29 |
262643_at |
AT1G62770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.201 |
0.217 |
| 30 |
264561_at |
AT1G55810
|
UKL3, uridine kinase-like 3 |
0.199 |
-0.005 |
| 31 |
249847_at |
AT5G23210
|
SCPL34, serine carboxypeptidase-like 34 |
0.198 |
0.149 |
| 32 |
264551_at |
AT1G09460
|
[Carbohydrate-binding X8 domain superfamily protein] |
0.197 |
0.006 |
| 33 |
250936_at |
AT5G03120
|
unknown |
0.197 |
-0.000 |
| 34 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
0.197 |
-0.164 |
| 35 |
259103_at |
AT3G11690
|
unknown |
0.195 |
0.069 |
| 36 |
266984_at |
AT2G39570
|
ACR9, ACT domain repeats 9 |
0.194 |
0.014 |
| 37 |
246018_at |
AT5G10695
|
unknown |
0.192 |
0.003 |
| 38 |
258227_at |
AT3G15620
|
UVR3, UV REPAIR DEFECTIVE 3 |
0.190 |
-0.058 |
| 39 |
261851_at |
AT1G50460
|
ATHKL1, HKL1, hexokinase-like 1 |
0.190 |
-0.023 |
| 40 |
262552_at |
AT1G31350
|
KUF1, KAR-UP F-box 1 |
0.181 |
-0.024 |
| 41 |
267246_at |
AT2G30250
|
ATWRKY25, WRKY25, WRKY DNA-binding protein 25 |
0.178 |
0.194 |
| 42 |
265771_at |
AT2G48030
|
[DNAse I-like superfamily protein] |
0.177 |
0.063 |
| 43 |
255625_at |
AT4G01120
|
ATBZIP54, BASIC REGION/LEUCINE ZIPPER MOTIF 5, GBF2, G-box binding factor 2 |
0.176 |
0.061 |
| 44 |
253571_at |
AT4G31000
|
[Calmodulin-binding protein] |
0.176 |
0.036 |
| 45 |
257990_at |
AT3G19860
|
bHLH121, basic Helix-Loop-Helix 121 |
0.175 |
0.061 |
| 46 |
262910_at |
AT1G59710
|
unknown |
0.173 |
-0.034 |
| 47 |
252991_at |
AT4G38470
|
STY46, serine/threonine/tyrosine kinase 46 |
0.173 |
0.103 |
| 48 |
247128_at |
AT5G66110
|
HIPP27, heavy metal associated isoprenylated plant protein 27 |
0.169 |
0.026 |
| 49 |
248744_at |
AT5G48250
|
BBX8, B-box domain protein 8 |
0.169 |
-0.017 |
| 50 |
258608_at |
AT3G03020
|
unknown |
0.167 |
0.108 |
| 51 |
250664_at |
AT5G07080
|
[HXXXD-type acyl-transferase family protein] |
0.165 |
0.006 |
| 52 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
0.165 |
0.006 |
| 53 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
0.165 |
0.221 |
| 54 |
246396_at |
AT1G58180
|
ATBCA6, A. THALIANA BETA CARBONIC ANHYDRASE 6, BCA6, beta carbonic anhydrase 6 |
0.164 |
0.071 |
| 55 |
260741_at |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
0.163 |
0.029 |
| 56 |
263460_at |
AT2G31810
|
[ACT domain-containing small subunit of acetolactate synthase protein] |
0.162 |
0.017 |
| 57 |
253802_at |
AT4G28180
|
unknown |
0.162 |
0.070 |
| 58 |
257654_at |
AT3G13310
|
DJC66, DNA J protein C66 |
0.162 |
-0.017 |
| 59 |
256697_at |
AT3G20660
|
AtOCT4, organic cation/carnitine transporter4, OCT4, organic cation/carnitine transporter4 |
0.161 |
0.046 |
| 60 |
255506_at |
AT4G02130
|
GATL6, galacturonosyltransferase-like 6, LGT10 |
0.160 |
-0.029 |
| 61 |
264815_at |
AT1G03620
|
[ELMO/CED-12 family protein] |
0.159 |
0.029 |
| 62 |
261211_at |
AT1G12780
|
ATUGE1, A. THALIANA UDP-GLC 4-EPIMERASE 1, UGE1, UDP-D-glucose/UDP-D-galactose 4-epimerase 1 |
0.159 |
0.094 |
| 63 |
246843_at |
AT5G26740
|
unknown |
0.158 |
0.004 |
| 64 |
254197_at |
AT4G24040
|
ATTRE1, TRE1, trehalase 1 |
0.158 |
0.018 |
| 65 |
248179_at |
AT5G54380
|
THE1, THESEUS1 |
0.158 |
0.011 |
| 66 |
247266_at |
AT5G64570
|
ATBXL4, ARABIDOPSIS THALIANA BETA-D-XYLOSIDASE 4, XYL4, beta-D-xylosidase 4 |
0.158 |
0.039 |
| 67 |
253829_at |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
0.157 |
0.092 |
| 68 |
260536_at |
AT2G43400
|
ETFQO, electron-transfer flavoprotein:ubiquinone oxidoreductase |
0.157 |
-0.018 |
| 69 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
0.156 |
0.045 |
| 70 |
248193_at |
AT5G54080
|
AtHGO, HGO, homogentisate 1,2-dioxygenase |
0.156 |
0.068 |
| 71 |
250032_at |
AT5G18170
|
GDH1, glutamate dehydrogenase 1 |
0.156 |
0.043 |
| 72 |
267279_at |
AT2G19460
|
unknown |
0.156 |
0.075 |
| 73 |
248796_at |
AT5G47180
|
[Plant VAMP (vesicle-associated membrane protein) family protein] |
0.155 |
0.003 |
| 74 |
252173_at |
AT3G50650
|
[GRAS family transcription factor] |
0.152 |
0.108 |
| 75 |
247123_at |
AT5G66050
|
[Wound-responsive family protein] |
0.150 |
0.065 |
| 76 |
251791_at |
AT3G55500
|
ATEXP16, ATEXPA16, expansin A16, ATHEXP ALPHA 1.7, EXP16, EXPANSIN 16, EXPA16, expansin A16 |
0.149 |
0.114 |
| 77 |
247954_at |
AT5G56870
|
BGAL4, beta-galactosidase 4 |
0.149 |
0.092 |
| 78 |
264763_at |
AT1G61450
|
unknown |
0.149 |
-0.026 |
| 79 |
260279_at |
AT1G80420
|
ATXRCC1, XRCC1, homolog of X-ray repair cross complementing 1 |
0.149 |
0.092 |
| 80 |
260988_at |
AT1G53570
|
MAP3KA, mitogen-activated protein kinase kinase kinase 3, MAPKKK3, MAP KINASE KINASE KINASE 3 |
0.148 |
-0.034 |
| 81 |
262525_at |
AT1G17060
|
CHI2, CHIBI 2, CYP72C1, cytochrome p450 72c1, SHK1, SHRINK 1, SOB7, SUPPRESSOR OF PHYB-4 7 |
0.148 |
0.062 |
| 82 |
262060_at |
AT1G80170
|
[Pectin lyase-like superfamily protein] |
0.147 |
0.023 |
| 83 |
247183_at |
AT5G65440
|
unknown |
0.147 |
0.034 |
| 84 |
267628_at |
AT2G42280
|
AKS3, ABA-responsive kinase substrate 3, FBH4, FLOWERING BHLH 4 |
0.147 |
-0.024 |
| 85 |
250809_at |
AT5G05140
|
[Transcription elongation factor (TFIIS) family protein] |
0.146 |
-0.070 |
| 86 |
264001_at |
AT2G22420
|
[Peroxidase superfamily protein] |
0.145 |
-0.024 |
| 87 |
261253_at |
AT1G05840
|
[Eukaryotic aspartyl protease family protein] |
0.142 |
0.000 |
| 88 |
247835_at |
AT5G57910
|
unknown |
0.141 |
0.196 |
| 89 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
0.141 |
0.279 |
| 90 |
258527_at |
AT3G06850
|
BCE2, DIN3, DARK INDUCIBLE 3, LTA1 |
0.140 |
-0.011 |
| 91 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.381 |
-0.177 |
| 92 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
-0.363 |
-0.549 |
| 93 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
-0.342 |
0.039 |
| 94 |
255284_at |
AT4G04610
|
APR, APR1, APS reductase 1, ATAPR1, PRH19, PAPS REDUCTASE HOMOLOG 19 |
-0.320 |
-0.062 |
| 95 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
-0.318 |
-0.143 |
| 96 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
-0.307 |
-0.071 |
| 97 |
250561_at |
AT5G08030
|
AtGDPD6, GDPD6, glycerophosphodiester phosphodiesterase 6 |
-0.306 |
-0.324 |
| 98 |
261177_at |
AT1G04770
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.303 |
-0.132 |
| 99 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.298 |
-0.469 |
| 100 |
263739_at |
AT2G21320
|
BBX18, B-box domain protein 18 |
-0.297 |
-0.265 |
| 101 |
259348_at |
AT3G03770
|
[Leucine-rich repeat protein kinase family protein] |
-0.293 |
-0.063 |
| 102 |
251058_at |
AT5G01790
|
unknown |
-0.291 |
-0.162 |
| 103 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
-0.290 |
-0.391 |
| 104 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
-0.284 |
-0.149 |
| 105 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.279 |
-0.104 |
| 106 |
245448_at |
AT4G16860
|
RPP4, recognition of peronospora parasitica 4 |
-0.278 |
-0.050 |
| 107 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
-0.275 |
0.122 |
| 108 |
265121_at |
AT1G62560
|
FMO GS-OX3, flavin-monooxygenase glucosinolate S-oxygenase 3 |
-0.272 |
-0.003 |
| 109 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
-0.267 |
-0.117 |
| 110 |
260955_at |
AT1G06000
|
[UDP-Glycosyltransferase superfamily protein] |
-0.267 |
-0.149 |
| 111 |
262935_at |
AT1G79410
|
AtOCT5, organic cation/carnitine transporter5, OCT5, organic cation/carnitine transporter5 |
-0.257 |
-0.039 |
| 112 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.247 |
-0.019 |
| 113 |
251024_at |
AT5G02180
|
[Transmembrane amino acid transporter family protein] |
-0.247 |
-0.096 |
| 114 |
252482_at |
AT3G46670
|
UGT76E11, UDP-glucosyl transferase 76E11 |
-0.243 |
-0.144 |
| 115 |
256603_at |
AT3G28270
|
unknown |
-0.237 |
0.043 |
| 116 |
245277_at |
AT4G15550
|
IAGLU, indole-3-acetate beta-D-glucosyltransferase |
-0.234 |
-0.114 |
| 117 |
249059_at |
AT5G44530
|
[Subtilase family protein] |
-0.234 |
-0.071 |
| 118 |
245676_at |
AT1G56670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.227 |
-0.034 |
| 119 |
263452_at |
AT2G22190
|
TPPE, trehalose-6-phosphate phosphatase E |
-0.222 |
-0.079 |
| 120 |
248028_at |
AT5G55620
|
unknown |
-0.222 |
-0.136 |
| 121 |
251060_at |
AT5G01820
|
ATCIPK14, ATSR1, serine/threonine protein kinase 1, CIPK14, CBL-INTERACTING PROTEIN KINASE 14, PKS24, SOS2-like protein kinase 24, SnRK3.15, SNF1-RELATED PROTEIN KINASE 3.15, SR1, serine/threonine protein kinase 1 |
-0.219 |
-0.047 |
| 122 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
-0.219 |
-0.042 |
| 123 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.217 |
0.214 |
| 124 |
251309_at |
AT3G61220
|
SDR1, short-chain dehydrogenase/reductase 1 |
-0.214 |
-0.129 |
| 125 |
246998_at |
AT5G67370
|
CGLD27, CONSERVED IN THE GREEN LINEAGE AND DIATOMS 27 |
-0.201 |
-0.233 |
| 126 |
258181_at |
AT3G21670
|
AtNPF6.4, NPF6.4, NRT1/ PTR family 6.4 |
-0.201 |
-0.176 |
| 127 |
255502_at |
AT4G02410
|
AtLPK1, LecRK-IV.3, L-type lectin receptor kinase IV.3, LPK1, lectin-like protein kinase 1 |
-0.200 |
-0.082 |
| 128 |
252911_at |
AT4G39510
|
CYP96A12, cytochrome P450, family 96, subfamily A, polypeptide 12 |
-0.199 |
0.003 |
| 129 |
250985_at |
AT5G02830
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.198 |
-0.133 |
| 130 |
259736_at |
AT1G64390
|
AtGH9C2, glycosyl hydrolase 9C2, GH9C2, glycosyl hydrolase 9C2 |
-0.197 |
0.003 |
| 131 |
246700_at |
AT5G28030
|
DES1, L-cysteine desulfhydrase 1 |
-0.193 |
-0.084 |
| 132 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
-0.192 |
-0.016 |
| 133 |
263252_at |
AT2G31380
|
BBX25, B-box domain protein 25, STH, salt tolerance homologue |
-0.192 |
-0.116 |
| 134 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
-0.191 |
-0.026 |
| 135 |
264745_at |
AT1G62180
|
5'adenylylphosphosulfate reductase 2 |
-0.186 |
-0.179 |
| 136 |
267538_at |
AT2G41870
|
[Remorin family protein] |
-0.184 |
0.073 |
| 137 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
-0.184 |
-0.435 |
| 138 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
-0.181 |
-0.283 |
| 139 |
260696_at |
AT1G32520
|
unknown |
-0.178 |
-0.134 |
| 140 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.177 |
-0.264 |
| 141 |
257272_at |
AT3G28130
|
UMAMIT44, Usually multiple acids move in and out Transporters 44 |
-0.172 |
0.028 |
| 142 |
261804_at |
AT1G30530
|
UGT78D1, UDP-glucosyl transferase 78D1 |
-0.171 |
-0.040 |
| 143 |
251005_at |
AT5G02590
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.169 |
-0.181 |
| 144 |
249774_at |
AT5G24150
|
SQE5, SQUALENE MONOOXYGENASE 5, SQP1 |
-0.168 |
-0.175 |
| 145 |
261319_at |
AT1G53090
|
SPA4, SPA1-related 4 |
-0.167 |
-0.113 |
| 146 |
250892_at |
AT5G03760
|
ATCSLA09, ATCSLA9, CSLA09, CSLA9, CELLULOSE SYNTHASE LIKE A9, RAT4, RESISTANT TO AGROBACTERIUM TRANSFORMATION 4 |
-0.164 |
-0.020 |
| 147 |
255032_at |
AT4G09500
|
[UDP-Glycosyltransferase superfamily protein] |
-0.162 |
-0.168 |
| 148 |
255278_at |
AT4G04940
|
[transducin family protein / WD-40 repeat family protein] |
-0.158 |
-0.125 |
| 149 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
-0.157 |
-0.019 |
| 150 |
262526_at |
AT1G17050
|
AtSPS2, SPS2, solanesyl diphosphate synthase 2 |
-0.157 |
-0.175 |
| 151 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
-0.156 |
-0.052 |
| 152 |
245965_at |
AT5G19730
|
[Pectin lyase-like superfamily protein] |
-0.156 |
-0.097 |
| 153 |
256673_at |
AT3G52370
|
FLA15, FASCICLIN-like arabinogalactan protein 15 precursor |
-0.154 |
-0.001 |
| 154 |
248961_at |
AT5G45650
|
[subtilase family protein] |
-0.153 |
-0.008 |
| 155 |
245936_at |
AT5G19850
|
[alpha/beta-Hydrolases superfamily protein] |
-0.153 |
-0.068 |
| 156 |
252679_at |
AT3G44260
|
AtCAF1a, CCR4- associated factor 1a, CAF1a, CCR4- associated factor 1a |
-0.152 |
-0.182 |
| 157 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
-0.150 |
-0.044 |
| 158 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
-0.148 |
0.070 |
| 159 |
246468_at |
AT5G17050
|
UGT78D2, UDP-glucosyl transferase 78D2 |
-0.147 |
-0.075 |
| 160 |
260601_at |
AT1G55910
|
ZIP11, zinc transporter 11 precursor |
-0.145 |
0.063 |
| 161 |
246249_at |
AT4G36680
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.144 |
-0.079 |
| 162 |
255694_at |
AT4G00050
|
UNE10, unfertilized embryo sac 10 |
-0.143 |
-0.162 |
| 163 |
257916_at |
AT3G23210
|
bHLH34, basic Helix-Loop-Helix 34 |
-0.143 |
-0.058 |
| 164 |
263777_at |
AT2G46450
|
ATCNGC12, cyclic nucleotide-gated channel 12, CNGC12, cyclic nucleotide-gated channel 12 |
-0.142 |
0.059 |
| 165 |
247323_at |
AT5G64170
|
LNK1, night light-inducible and clock-regulated 1 |
-0.142 |
-0.018 |
| 166 |
247351_at |
AT5G63790
|
ANAC102, NAC domain containing protein 102, NAC102, NAC domain containing protein 102 |
-0.142 |
-0.122 |
| 167 |
260676_at |
AT1G19450
|
[Major facilitator superfamily protein] |
-0.140 |
-0.091 |
| 168 |
246403_at |
AT1G57590
|
[Pectinacetylesterase family protein] |
-0.139 |
-0.012 |
| 169 |
247549_at |
AT5G61420
|
AtMYB28, HAG1, HIGH ALIPHATIC GLUCOSINOLATE 1, MYB28, myb domain protein 28, PMG1, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 1 |
-0.139 |
-0.020 |
| 170 |
247716_at |
AT5G59350
|
unknown |
-0.139 |
-0.022 |
| 171 |
251811_at |
AT3G54990
|
SMZ, SCHLAFMUTZE |
-0.138 |
-0.069 |
| 172 |
247118_at |
AT5G65890
|
ACR1, ACT domain repeat 1 |
-0.137 |
-0.023 |
| 173 |
253915_at |
AT4G27280
|
[Calcium-binding EF-hand family protein] |
-0.137 |
-0.315 |
| 174 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
-0.136 |
-0.484 |
| 175 |
260135_at |
AT1G66400
|
CML23, calmodulin like 23 |
-0.136 |
-0.164 |
| 176 |
252639_at |
AT3G44550
|
FAR5, fatty acid reductase 5 |
-0.135 |
-0.000 |
| 177 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
-0.135 |
-0.094 |
| 178 |
246125_at |
AT5G19875
|
unknown |
-0.134 |
-0.024 |
| 179 |
253922_at |
AT4G26850
|
VTC2, vitamin c defective 2 |
-0.134 |
-0.062 |