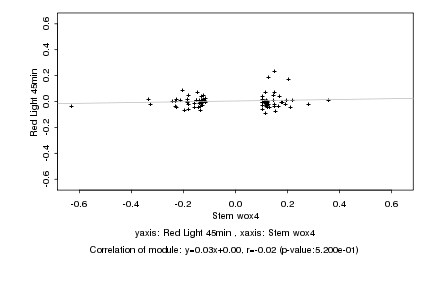
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Stem wox4 |
Log2 signal ratio
Red Light 45min |
| 1 |
257481_at |
AT1G08430
|
ALMT1, aluminum-activated malate transporter 1, ATALMT1, ARABIDOPSIS THALIANA ALUMINUM-ACTIVATED MALATE TRANSPORTER 1 |
0.355 |
0.013 |
| 2 |
249727_at |
AT5G35490
|
ATMRU1, ARABIDOPSIS MTO 1 RESPONDING UP 1, MRU1, MTO 1 RESPONDING UP 1 |
0.279 |
-0.022 |
| 3 |
247377_at |
AT5G63180
|
[Pectin lyase-like superfamily protein] |
0.217 |
0.011 |
| 4 |
252501_at |
AT3G46880
|
unknown |
0.211 |
-0.041 |
| 5 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
0.201 |
0.172 |
| 6 |
246900_at |
AT5G25620
|
AtYUC6, YUC6, YUCCA6 |
0.196 |
0.011 |
| 7 |
250661_at |
AT5G07030
|
[Eukaryotic aspartyl protease family protein] |
0.191 |
-0.019 |
| 8 |
247850_at |
AT5G58150
|
[Leucine-rich repeat protein kinase family protein] |
0.178 |
-0.005 |
| 9 |
262902_x_at |
AT1G59930
|
[MADS-box family protein] |
0.177 |
-0.003 |
| 10 |
250471_at |
AT5G10170
|
ATMIPS3, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 3, MIPS3, myo-inositol-1-phosphate synthase 3 |
0.166 |
0.041 |
| 11 |
253966_at |
AT4G26520
|
AtFBA7, FBA7, fructose-bisphosphate aldolase 7 |
0.165 |
-0.031 |
| 12 |
246107_at |
AT5G28700
|
[pseudogene] |
0.152 |
-0.076 |
| 13 |
249726_at |
AT5G35480
|
unknown |
0.150 |
-0.031 |
| 14 |
250574_at |
AT5G08230
|
[Tudor/PWWP/MBT domain-containing protein] |
0.149 |
-0.018 |
| 15 |
248230_at |
AT5G53830
|
[VQ motif-containing protein] |
0.149 |
0.076 |
| 16 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
0.147 |
0.232 |
| 17 |
247116_at |
AT5G65970
|
ATMLO10, MILDEW RESISTANCE LOCUS O 10, MLO10, MILDEW RESISTANCE LOCUS O 10 |
0.144 |
0.048 |
| 18 |
248976_at |
AT5G44930
|
ARAD2, ARABINAN DEFICIENT 2 |
0.143 |
0.011 |
| 19 |
254069_at |
AT4G25434
|
ATNUDT10, nudix hydrolase homolog 10, NUDT10, nudix hydrolase homolog 10 |
0.130 |
-0.046 |
| 20 |
247796_at |
AT5G58782
|
AtcPT6, cPT6, cis-prenyltransferase 6 |
0.126 |
-0.022 |
| 21 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
0.124 |
0.193 |
| 22 |
263883_at |
AT2G21830
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.123 |
0.002 |
| 23 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
0.122 |
-0.001 |
| 24 |
258751_at |
AT3G05890
|
RCI2B, RARE-COLD-INDUCIBLE 2B |
0.121 |
-0.016 |
| 25 |
259478_at |
AT1G18980
|
[RmlC-like cupins superfamily protein] |
0.120 |
-0.042 |
| 26 |
263549_at |
AT2G21650
|
ATRL2, ARABIDOPSIS RAD-LIKE 2, MEE3, MATERNAL EFFECT EMBRYO ARREST 3, RSM1, RADIALIS-LIKE SANT/MYB 1 |
0.119 |
-0.030 |
| 27 |
251246_at |
AT3G62100
|
IAA30, indole-3-acetic acid inducible 30 |
0.119 |
-0.033 |
| 28 |
251895_at |
AT3G54420
|
ATCHITIV, CHITINASE CLASS IV, ATEP3, homolog of carrot EP3-3 chitinase, CHIV, EP3, homolog of carrot EP3-3 chitinase |
0.117 |
-0.019 |
| 29 |
262630_at |
AT1G06520
|
ATGPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1, GPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1, sn-2-GPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1 |
0.117 |
-0.002 |
| 30 |
260801_at |
AT1G78430
|
RIP4, ROP interactive partner 4 |
0.116 |
0.013 |
| 31 |
249570_at |
AT5G38080
|
unknown |
0.114 |
-0.086 |
| 32 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
0.113 |
0.071 |
| 33 |
252834_at |
AT4G40070
|
[RING/U-box superfamily protein] |
0.112 |
-0.014 |
| 34 |
263788_at |
AT2G24580
|
[FAD-dependent oxidoreductase family protein] |
0.112 |
0.006 |
| 35 |
255157_at |
AT4G07790
|
[hypothetical protein] |
0.111 |
0.003 |
| 36 |
245637_at |
AT1G25230
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
0.110 |
-0.006 |
| 37 |
264962_at |
AT1G77110
|
PIN6, PIN-FORMED 6 |
0.104 |
-0.027 |
| 38 |
256976_at |
AT3G21020
|
[copia-like retrotransposon family, has a 5.4e-14 P-value blast match to gb|AAO73529.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
0.103 |
0.039 |
| 39 |
254303_at |
AT4G22830
|
unknown |
0.103 |
-0.007 |
| 40 |
253372_at |
AT4G33220
|
ATPME44, A. THALIANA PECTIN METHYLESTERASE 44, PME44, pectin methylesterase 44 |
0.103 |
-0.003 |
| 41 |
261816_at |
AT1G08135
|
ATCHX6B, CHX6B, cation/H+ exchanger 6B |
0.102 |
-0.059 |
| 42 |
262827_at |
AT1G13100
|
CYP71B29, cytochrome P450, family 71, subfamily B, polypeptide 29 |
0.102 |
0.018 |
| 43 |
263161_at |
AT1G54020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.633 |
-0.036 |
| 44 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
-0.335 |
0.019 |
| 45 |
266663_at |
AT2G25790
|
SKM1, STERILITY-REGULATING KINASE MEMBER 1 |
-0.328 |
-0.019 |
| 46 |
249817_at |
AT5G23820
|
ML3, MD2-related lipid recognition 3 |
-0.243 |
0.006 |
| 47 |
259385_at |
AT1G13470
|
unknown |
-0.233 |
0.004 |
| 48 |
254562_at |
AT4G19230
|
CYP707A1, cytochrome P450, family 707, subfamily A, polypeptide 1 |
-0.233 |
-0.032 |
| 49 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
-0.230 |
-0.039 |
| 50 |
248963_at |
AT5G45700
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.228 |
0.018 |
| 51 |
256389_at |
AT3G06220
|
[AP2/B3-like transcriptional factor family protein] |
-0.212 |
0.012 |
| 52 |
247425_at |
AT5G62550
|
unknown |
-0.205 |
0.088 |
| 53 |
266669_at |
AT2G29750
|
UGT71C1, UDP-glucosyl transferase 71C1 |
-0.197 |
-0.062 |
| 54 |
266555_at |
AT2G46270
|
GBF3, G-box binding factor 3 |
-0.188 |
0.005 |
| 55 |
267555_at |
AT2G32765
|
ATSUMO5, SUM5, SUMO 5, SUMO5, small ubiquitinrelated modifier 5 |
-0.188 |
0.022 |
| 56 |
266503_at |
AT2G47780
|
[Rubber elongation factor protein (REF)] |
-0.187 |
-0.000 |
| 57 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
-0.182 |
-0.054 |
| 58 |
254361_at |
AT4G22212
|
[Arabidopsis defensin-like protein] |
-0.181 |
-0.017 |
| 59 |
247696_at |
AT5G59780
|
ATMYB59, MYB DOMAIN PROTEIN 59, ATMYB59-1, ATMYB59-2, ATMYB59-3, MYB59, myb domain protein 59 |
-0.181 |
0.049 |
| 60 |
259753_at |
AT1G71050
|
HIPP20, heavy metal associated isoprenylated plant protein 20 |
-0.159 |
-0.013 |
| 61 |
259616_at |
AT1G47960
|
ATC/VIF1, CELL WALL / VACUOLAR INHIBITOR OF FRUCTOSIDASE 1, C/VIF1, cell wall / vacuolar inhibitor of fructosidase 1 |
-0.158 |
-0.043 |
| 62 |
260399_at |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
-0.151 |
0.015 |
| 63 |
266655_at |
AT2G25880
|
AtAUR2, ataurora2, AUR2, ataurora2 |
-0.150 |
0.008 |
| 64 |
245662_at |
AT1G28190
|
unknown |
-0.146 |
0.076 |
| 65 |
253684_at |
AT4G29690
|
[Alkaline-phosphatase-like family protein] |
-0.143 |
-0.040 |
| 66 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
-0.139 |
0.014 |
| 67 |
265414_at |
AT2G16660
|
[Major facilitator superfamily protein] |
-0.139 |
-0.015 |
| 68 |
252387_at |
AT3G47800
|
[Galactose mutarotase-like superfamily protein] |
-0.138 |
-0.062 |
| 69 |
255265_at |
AT4G05190
|
ATK5, kinesin 5 |
-0.137 |
-0.000 |
| 70 |
256081_at |
AT1G20700
|
ATWOX14, WUSCHEL RELATED HOMEOBOX 14, WOX14, WUSCHEL related homeobox 14 |
-0.137 |
-0.035 |
| 71 |
247603_at |
AT5G60930
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.134 |
0.010 |
| 72 |
248660_at |
AT5G48650
|
[Nuclear transport factor 2 (NTF2) family protein with RNA binding (RRM-RBD-RNP motifs) domain] |
-0.133 |
0.040 |
| 73 |
259302_at |
AT3G05120
|
ATGID1A, GA INSENSITIVE DWARF1A, GID1A, GA INSENSITIVE DWARF1A |
-0.131 |
-0.009 |
| 74 |
248413_at |
AT5G51600
|
ATMAP65-3, ARABIDOPSIS THALIANA MICROTUBULE-ASSOCIATED PROTEIN 65-3, MAP65-3, MICROTUBULE-ASSOCIATED PROTEIN 65-3, PLE, PLEIADE |
-0.131 |
0.008 |
| 75 |
265452_at |
AT2G46510
|
AIB, ABA-inducible BHLH-type transcription factor, ATAIB, ABA-inducible BHLH-type transcription factor, JAM1, JA-associated MYC2-like 1 |
-0.129 |
-0.007 |
| 76 |
249379_at |
AT5G40460
|
unknown |
-0.128 |
-0.028 |
| 77 |
252684_at |
AT3G44400
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.128 |
0.012 |
| 78 |
246929_at |
AT5G25210
|
unknown |
-0.126 |
0.049 |
| 79 |
247962_at |
AT5G56580
|
ANQ1, ARABIDOPSIS NQK1, ATMKK6, ARABIDOPSIS THALIANA MAP KINASE KINASE 6, MKK6, MAP kinase kinase 6 |
-0.125 |
0.018 |
| 80 |
250785_at |
AT5G05510
|
[Mad3/BUB1 homology region 1] |
-0.122 |
0.013 |
| 81 |
253771_at |
AT4G28430
|
[Reticulon family protein] |
-0.118 |
0.002 |
| 82 |
257216_at |
AT3G14990
|
AtDJ1A, DJ-1 homolog A, DJ-1a, DJ1A, DJ-1 homolog A |
-0.116 |
-0.007 |
| 83 |
245343_at |
AT4G15830
|
[ARM repeat superfamily protein] |
-0.116 |
0.028 |