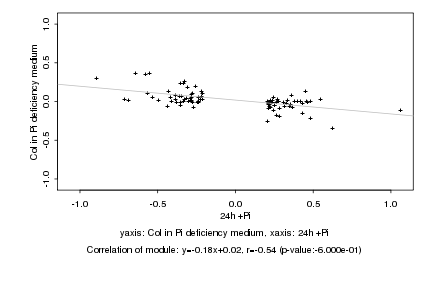
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
24h +Pi |
Log2 signal ratio
Col in Pi deficiency medium |
| 1 |
254550_at |
AT4G19690
|
ATIRT1, ARABIDOPSIS IRON-REGULATED TRANSPORTER 1, IRT1, iron-regulated transporter 1 |
1.059 |
-0.111 |
| 2 |
257720_at |
AT3G18450
|
[PLAC8 family protein] |
0.622 |
-0.337 |
| 3 |
264892_at |
AT1G23160
|
[Auxin-responsive GH3 family protein] |
0.544 |
0.027 |
| 4 |
267457_at |
AT2G33790
|
AGP30, arabinogalactan protein 30, ATAGP30 |
0.482 |
0.013 |
| 5 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.480 |
-0.217 |
| 6 |
254313_at |
AT4G22460
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.458 |
-0.010 |
| 7 |
248343_at |
AT5G52260
|
AtMYB19, myb domain protein 19, MYB19, myb domain protein 19 |
0.456 |
0.002 |
| 8 |
253179_at |
AT4G35200
|
unknown |
0.448 |
0.132 |
| 9 |
261466_at |
AT1G07690
|
unknown |
0.427 |
-0.015 |
| 10 |
264470_at |
AT1G67110
|
CYP735A2, cytochrome P450, family 735, subfamily A, polypeptide 2 |
0.426 |
-0.143 |
| 11 |
255911_at |
AT1G66930
|
[Protein kinase superfamily protein] |
0.413 |
0.006 |
| 12 |
245571_at |
AT4G14695
|
[Uncharacterised protein family (UPF0041)] |
0.396 |
0.002 |
| 13 |
256563_at |
AT3G29780
|
RALFL27, ralf-like 27 |
0.374 |
0.011 |
| 14 |
261550_at |
AT1G63450
|
RHS8, root hair specific 8, XUT1, XYLOGLUCAN-SPECIFIC GALACTURONOSYLTRANSFERASE 1 |
0.364 |
-0.072 |
| 15 |
266477_at |
AT2G31085
|
AtCLE6, CLE6, CLAVATA3/ESR-RELATED 6 |
0.355 |
0.088 |
| 16 |
260846_at |
AT1G17300
|
unknown |
0.352 |
-0.028 |
| 17 |
253309_at |
AT4G33790
|
CER4, ECERIFERUM 4, FAR3, FATTY ACID REDUCTASE 3, G7 |
0.346 |
-0.052 |
| 18 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
0.334 |
0.019 |
| 19 |
252502_at |
AT3G46900
|
COPT2, copper transporter 2 |
0.324 |
-0.013 |
| 20 |
260122_at |
AT1G33900
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.314 |
-0.053 |
| 21 |
248898_at |
AT5G46370
|
ATKCO2, CA2+ ACTIVATED OUTWARD RECTIFYING K+ CHANNEL 2, ATTPK2, TANDEM PORE K+ CHANNEL 2, KCO2, Ca2+ activated outward rectifying K+ channel 2 |
0.308 |
-0.008 |
| 22 |
266066_at |
AT2G18800
|
ATXTH21, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 21, XTH21, xyloglucan endotransglucosylase/hydrolase 21 |
0.283 |
-0.186 |
| 23 |
249768_at |
AT5G24100
|
[Leucine-rich repeat protein kinase family protein] |
0.281 |
-0.090 |
| 24 |
263765_at |
AT2G21540
|
ATSFH3, SEC14-LIKE 3, SFH3, SEC14-like 3 |
0.273 |
0.011 |
| 25 |
265048_at |
AT1G52050
|
[Mannose-binding lectin superfamily protein] |
0.271 |
0.001 |
| 26 |
266688_at |
AT2G19660
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.266 |
0.001 |
| 27 |
248916_at |
AT5G45840
|
[Leucine-rich repeat protein kinase family protein] |
0.265 |
0.031 |
| 28 |
265049_at |
AT1G52060
|
[Mannose-binding lectin superfamily protein] |
0.263 |
-0.009 |
| 29 |
246108_at |
AT5G28630
|
[glycine-rich protein] |
0.263 |
-0.175 |
| 30 |
267409_at |
AT2G34910
|
unknown |
0.262 |
-0.003 |
| 31 |
258143_at |
AT3G18170
|
[Glycosyltransferase family 61 protein] |
0.247 |
-0.045 |
| 32 |
254774_at |
AT4G13440
|
[Calcium-binding EF-hand family protein] |
0.247 |
-0.047 |
| 33 |
250157_at |
AT5G15180
|
[Peroxidase superfamily protein] |
0.244 |
-0.115 |
| 34 |
247474_at |
AT5G62280
|
unknown |
0.240 |
0.057 |
| 35 |
247991_at |
AT5G56320
|
ATEXP14, ATEXPA14, expansin A14, ATHEXP ALPHA 1.5, EXP14, EXPANSIN 14, EXPA14, expansin A14 |
0.237 |
-0.012 |
| 36 |
261226_at |
AT1G20190
|
ATEXP11, ATEXPA11, expansin 11, ATHEXP ALPHA 1.14, EXP11, EXPANSIN 11, EXPA11, expansin 11 |
0.236 |
0.026 |
| 37 |
264988_at |
AT1G27140
|
ATGSTU14, glutathione S-transferase tau 14, GST13, GLUTATHIONE S-TRANSFERASE 13, GSTU14, glutathione S-transferase tau 14 |
0.229 |
-0.009 |
| 38 |
250658_at |
AT5G07040
|
[RING/U-box superfamily protein] |
0.222 |
0.021 |
| 39 |
247582_at |
AT5G60760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.220 |
-0.074 |
| 40 |
263836_at |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
0.217 |
-0.060 |
| 41 |
266083_at |
AT2G37820
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.216 |
-0.007 |
| 42 |
244936_at |
ATCG01100
|
NDHA |
0.213 |
-0.030 |
| 43 |
251232_at |
AT3G62780
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.211 |
-0.030 |
| 44 |
261745_at |
AT1G08500
|
AtENODL18, ENODL18, early nodulin-like protein 18 |
0.210 |
-0.018 |
| 45 |
262978_at |
AT1G75780
|
TUB1, tubulin beta-1 chain |
0.210 |
-0.008 |
| 46 |
259276_at |
AT3G01190
|
[Peroxidase superfamily protein] |
0.209 |
-0.083 |
| 47 |
245979_at |
AT5G13150
|
ATEXO70C1, exocyst subunit exo70 family protein C1, EXO70C1, exocyst subunit exo70 family protein C1 |
0.209 |
-0.007 |
| 48 |
254912_at |
AT4G11230
|
[Riboflavin synthase-like superfamily protein] |
0.204 |
0.008 |
| 49 |
247685_at |
AT5G59680
|
[Leucine-rich repeat protein kinase family protein] |
0.204 |
-0.247 |
| 50 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
-0.894 |
0.304 |
| 51 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
-0.716 |
0.036 |
| 52 |
248545_at |
AT5G50260
|
CEP1, cysteine endopeptidase 1 |
-0.692 |
0.024 |
| 53 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
-0.643 |
0.366 |
| 54 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
-0.583 |
0.351 |
| 55 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
-0.566 |
0.109 |
| 56 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
-0.556 |
0.374 |
| 57 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
-0.535 |
0.056 |
| 58 |
248618_at |
AT5G49620
|
AtMYB78, myb domain protein 78, MYB78, myb domain protein 78 |
-0.499 |
0.020 |
| 59 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
-0.440 |
-0.054 |
| 60 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
-0.433 |
0.140 |
| 61 |
247052_at |
AT5G66700
|
ATHB53, ARABIDOPSIS THALIANA HOMEOBOX 53, HB-8, HOMEOBOX-8, HB53, homeobox 53 |
-0.420 |
0.055 |
| 62 |
247326_at |
AT5G64110
|
[Peroxidase superfamily protein] |
-0.413 |
0.004 |
| 63 |
260386_at |
AT1G74010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.390 |
0.039 |
| 64 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
-0.388 |
0.089 |
| 65 |
258516_at |
AT3G06490
|
AtMYB108, myb domain protein 108, BOS1, BOTRYTIS-SUSCEPTIBLE1, MYB108, myb domain protein 108 |
-0.382 |
-0.012 |
| 66 |
245463_at |
AT4G17030
|
AT-EXPR, ATEXLB1, expansin-like B1, ATEXPR1, ATHEXP BETA 3.1, EXLB1, expansin-like B1, EXPR |
-0.364 |
0.077 |
| 67 |
265769_at |
AT2G48090
|
unknown |
-0.358 |
-0.007 |
| 68 |
258856_at |
AT3G02040
|
AtGDPD1, GDPD1, Glycerophosphodiester phosphodiesterase 1, SRG3, senescence-related gene 3 |
-0.357 |
0.241 |
| 69 |
266974_at |
AT2G39370
|
MAKR4, MEMBRANE-ASSOCIATED KINASE REGULATOR 4 |
-0.355 |
-0.040 |
| 70 |
250646_at |
AT5G06720
|
ATPA2, peroxidase 2, AtPRX53, PA2, peroxidase 2, PRX53, peroxidase 53 |
-0.350 |
0.074 |
| 71 |
266132_at |
AT2G45130
|
ATSPX3, ARABIDOPSIS THALIANA SPX DOMAIN GENE 3, SPX3, SPX domain gene 3 |
-0.339 |
0.008 |
| 72 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
-0.336 |
0.236 |
| 73 |
254764_at |
AT4G13250
|
NYC1, NON-YELLOW COLORING 1 |
-0.334 |
0.039 |
| 74 |
252414_at |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
-0.332 |
0.260 |
| 75 |
265411_at |
AT2G16630
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.320 |
0.044 |
| 76 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
-0.313 |
0.192 |
| 77 |
246000_at |
AT5G20820
|
SAUR76, SMALL AUXIN UPREGULATED RNA 76 |
-0.302 |
0.012 |
| 78 |
248912_at |
AT5G45670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.295 |
0.023 |
| 79 |
254034_at |
AT4G25960
|
ABCB2, ATP-binding cassette B2, PGP2, P-glycoprotein 2 |
-0.295 |
0.046 |
| 80 |
257697_at |
AT3G12700
|
NANA, NANA |
-0.288 |
0.056 |
| 81 |
249992_at |
AT5G18560
|
PUCHI |
-0.286 |
0.018 |
| 82 |
245904_at |
AT5G11110
|
ATSPS2F, sucrose phosphate synthase 2F, KNS2, KAONASHI 2, SPS1, SUCROSE PHOSPHATE SYNTHASE 1, SPS2F, sucrose phosphate synthase 2F, SPSA2, sucrose-phosphate synthase A2 |
-0.285 |
0.101 |
| 83 |
265025_at |
AT1G24575
|
unknown |
-0.284 |
0.012 |
| 84 |
246055_at |
AT5G08380
|
AGAL1, alpha-galactosidase 1, AtAGAL1, alpha-galactosidase 1 |
-0.278 |
-0.000 |
| 85 |
259173_at |
AT3G03640
|
BGLU25, beta glucosidase 25, GLUC |
-0.277 |
0.114 |
| 86 |
258209_at |
AT3G14060
|
unknown |
-0.276 |
-0.075 |
| 87 |
253657_at |
AT4G30110
|
ATHMA2, ARABIDOPSIS HEAVY METAL ATPASE 2, HMA2, heavy metal atpase 2 |
-0.260 |
0.202 |
| 88 |
253872_at |
AT4G27410
|
ANAC072, Arabidopsis NAC domain containing protein 72, RD26, RESPONSIVE TO DESICCATION 26 |
-0.249 |
-0.006 |
| 89 |
258887_at |
AT3G05630
|
PDLZ2, PHOSPHOLIPASE D ZETA 2, PLDP2, phospholipase D P2, PLDZETA2, PHOSPHOLIPASE D ZETA 2 |
-0.244 |
0.004 |
| 90 |
260933_at |
AT1G02470
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.241 |
-0.004 |
| 91 |
264130_at |
AT1G79160
|
unknown |
-0.241 |
0.011 |
| 92 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
-0.238 |
0.061 |
| 93 |
246228_at |
AT4G36430
|
[Peroxidase superfamily protein] |
-0.226 |
0.030 |
| 94 |
256927_at |
AT3G22550
|
unknown |
-0.221 |
0.074 |
| 95 |
249747_at |
AT5G24600
|
unknown |
-0.221 |
0.130 |
| 96 |
259656_at |
AT1G55200
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.218 |
0.029 |
| 97 |
256627_at |
AT3G19970
|
[alpha/beta-Hydrolases superfamily protein] |
-0.213 |
0.114 |