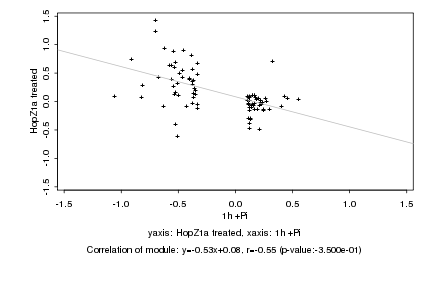
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
1h +Pi |
Log2 signal ratio
HopZ1a treated |
| 1 |
265683_at |
AT2G24400
|
SAUR38, SMALL AUXIN UPREGULATED RNA 38 |
0.544 |
0.053 |
| 2 |
250992_at |
AT5G02260
|
ATEXP9, ATEXPA9, expansin A9, ATHEXP ALPHA 1.10, EXP9, EXPA9, expansin A9 |
0.454 |
0.062 |
| 3 |
253515_at |
AT4G31320
|
SAUR37, SMALL AUXIN UPREGULATED RNA 37 |
0.422 |
0.101 |
| 4 |
255591_at |
AT4G01630
|
ATEXP17, EXPANSIN 17, ATEXPA17, expansin A17, ATHEXP ALPHA 1.13, EXPA17, expansin A17 |
0.402 |
-0.081 |
| 5 |
260276_at |
AT1G80450
|
[VQ motif-containing protein] |
0.321 |
0.717 |
| 6 |
263194_at |
AT1G36060
|
[Integrase-type DNA-binding superfamily protein] |
0.291 |
-0.127 |
| 7 |
255911_at |
AT1G66930
|
[Protein kinase superfamily protein] |
0.267 |
0.016 |
| 8 |
265350_at |
AT2G22620
|
[Rhamnogalacturonate lyase family protein] |
0.258 |
0.067 |
| 9 |
249709_at |
AT5G35670
|
iqd33, IQ-domain 33 |
0.242 |
-0.147 |
| 10 |
259994_at |
AT1G68130
|
AtIDD14, indeterminate(ID)-domain 14, IDD14, indeterminate(ID)-domain 14, IDD14alpha, IDD14beta |
0.237 |
-0.130 |
| 11 |
262864_at |
AT1G64920
|
[UDP-Glycosyltransferase superfamily protein] |
0.231 |
-0.014 |
| 12 |
247013_at |
AT5G67480
|
ATBT4, BT4, BTB and TAZ domain protein 4 |
0.226 |
-0.042 |
| 13 |
262981_at |
AT1G75590
|
SAUR52, SMALL AUXIN UPREGULATED RNA 52 |
0.218 |
0.025 |
| 14 |
248989_at |
AT5G45200
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.213 |
-0.012 |
| 15 |
265249_at |
AT2G01940
|
ATIDD15, ARABIDOPSIS THALIANA INDETERMINATE(ID)-DOMAIN 15, SGR5, SHOOT GRAVITROPISM 5 |
0.210 |
-0.063 |
| 16 |
266873_at |
AT2G44740
|
CYCP4;1, cyclin p4;1 |
0.206 |
-0.477 |
| 17 |
257353_at |
AT2G23050
|
MEL4, MAB4/ENP/NPY1-LIKE 4, NPY4, NAKED PINS IN YUC MUTANTS 4 |
0.201 |
0.057 |
| 18 |
248584_at |
AT5G49960
|
unknown |
0.187 |
-0.139 |
| 19 |
260395_at |
AT1G69780
|
ATHB13 |
0.180 |
0.046 |
| 20 |
261581_at |
AT1G01140
|
CIPK9, CBL-interacting protein kinase 9, PKS6, PROTEIN KINASE 6, SnRK3.12, SNF1-RELATED PROTEIN KINASE 3.12 |
0.168 |
0.083 |
| 21 |
253145_at |
AT4G35560
|
DAW1, DUO1-activated WD40 1 |
0.166 |
-0.032 |
| 22 |
260992_at |
AT1G12150
|
unknown |
0.165 |
0.114 |
| 23 |
260277_at |
AT1G80520
|
[Sterile alpha motif (SAM) domain-containing protein] |
0.164 |
0.113 |
| 24 |
257460_at |
AT1G75580
|
SAUR51, SMALL AUXIN UPREGULATED RNA 51 |
0.162 |
-0.134 |
| 25 |
265544_at |
AT2G28260
|
ATCNGC15, cyclic nucleotide-gated channel 15, CNGC15, cyclic nucleotide-gated channel 15 |
0.152 |
-0.064 |
| 26 |
264470_at |
AT1G67110
|
CYP735A2, cytochrome P450, family 735, subfamily A, polypeptide 2 |
0.148 |
-0.050 |
| 27 |
264127_at |
AT1G79250
|
AGC1.7, AGC kinase 1.7 |
0.142 |
0.110 |
| 28 |
255757_at |
AT4G00460
|
ATROPGEF3, ROPGEF3, ROP (rho of plants) guanine nucleotide exchange factor 3 |
0.141 |
0.122 |
| 29 |
246964_at |
AT5G24830
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.139 |
-0.055 |
| 30 |
260181_at |
AT1G70710
|
ATGH9B1, glycosyl hydrolase 9B1, CEL1, CELLULASE 1, GH9B1, glycosyl hydrolase 9B1 |
0.134 |
-0.101 |
| 31 |
249059_at |
AT5G44530
|
[Subtilase family protein] |
0.129 |
-0.304 |
| 32 |
248299_at |
AT5G53080
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.125 |
-0.297 |
| 33 |
265847_at |
AT2G35750
|
unknown |
0.120 |
-0.145 |
| 34 |
247877_at |
AT5G57740
|
XBAT32, XB3 ortholog 2 in Arabidopsis thaliana |
0.120 |
0.105 |
| 35 |
260883_at |
AT1G29270
|
unknown |
0.120 |
0.073 |
| 36 |
254803_at |
AT4G13100
|
[RING/U-box superfamily protein] |
0.120 |
-0.472 |
| 37 |
260494_at |
AT2G41820
|
PXC3, PXY/TDR-correlated 3 |
0.118 |
-0.371 |
| 38 |
249088_at |
AT5G44230
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.118 |
-0.092 |
| 39 |
247687_at |
AT5G59740
|
[UDP-N-acetylglucosamine (UAA) transporter family] |
0.115 |
0.026 |
| 40 |
261750_at |
AT1G76120
|
[Pseudouridine synthase family protein] |
0.113 |
-0.046 |
| 41 |
259174_at |
AT3G03690
|
UNE7, unfertilized embryo sac 7 |
0.112 |
0.087 |
| 42 |
263841_at |
AT2G36870
|
AtXTH32, XTH32, xyloglucan endotransglucosylase/hydrolase 32 |
0.110 |
-0.292 |
| 43 |
267011_at |
AT2G39230
|
LOJ, LATERAL ORGAN JUNCTION |
0.108 |
-0.022 |
| 44 |
266869_at |
AT2G44660
|
[ALG6, ALG8 glycosyltransferase family] |
0.104 |
0.091 |
| 45 |
259585_at |
AT1G28090
|
[Polynucleotide adenylyltransferase family protein] |
0.102 |
0.018 |
| 46 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
-1.064 |
0.095 |
| 47 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
-0.913 |
0.756 |
| 48 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
-0.829 |
0.082 |
| 49 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
-0.816 |
0.297 |
| 50 |
249197_at |
AT5G42380
|
CML37, calmodulin like 37 |
-0.706 |
1.442 |
| 51 |
259866_at |
AT1G76640
|
CML39, CALMODULIN LIKE 39 |
-0.702 |
1.236 |
| 52 |
251774_at |
AT3G55840
|
[Hs1pro-1 protein] |
-0.676 |
0.435 |
| 53 |
252997_at |
AT4G38400
|
ATEXLA2, expansin-like A2, ATEXPL2, ATHEXP BETA 2.2, EXLA2, expansin-like A2, EXPL2, EXPANSIN L2 |
-0.634 |
-0.074 |
| 54 |
262085_at |
AT1G56060
|
unknown |
-0.624 |
0.945 |
| 55 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
-0.580 |
0.646 |
| 56 |
247047_at |
AT5G66650
|
unknown |
-0.563 |
0.389 |
| 57 |
266010_at |
AT2G37430
|
ZAT11, zinc finger of Arabidopsis thaliana 11 |
-0.563 |
0.640 |
| 58 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
-0.548 |
0.270 |
| 59 |
253632_at |
AT4G30430
|
TET9, tetraspanin9 |
-0.547 |
0.887 |
| 60 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
-0.540 |
0.603 |
| 61 |
252070_at |
AT3G51680
|
AtSDR2, SDR2, short-chain dehydrogenase/reductase 2 |
-0.535 |
0.127 |
| 62 |
248789_at |
AT5G47440
|
unknown |
-0.530 |
-0.390 |
| 63 |
253485_at |
AT4G31800
|
ATWRKY18, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 18, WRKY18, WRKY DNA-binding protein 18 |
-0.528 |
0.172 |
| 64 |
247622_at |
AT5G60350
|
unknown |
-0.528 |
0.695 |
| 65 |
246200_at |
AT4G37240
|
unknown |
-0.510 |
-0.598 |
| 66 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
-0.508 |
0.331 |
| 67 |
262124_at |
AT1G59660
|
[Nucleoporin autopeptidase] |
-0.503 |
0.114 |
| 68 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
-0.496 |
0.507 |
| 69 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
-0.468 |
0.548 |
| 70 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
-0.465 |
0.436 |
| 71 |
247208_at |
AT5G64870
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
-0.458 |
0.900 |
| 72 |
252193_at |
AT3G50060
|
MYB77, myb domain protein 77 |
-0.437 |
-0.073 |
| 73 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
-0.408 |
0.419 |
| 74 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
-0.408 |
0.388 |
| 75 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
-0.392 |
0.814 |
| 76 |
259325_at |
AT3G05320
|
[O-fucosyltransferase family protein] |
-0.383 |
0.308 |
| 77 |
253643_at |
AT4G29780
|
unknown |
-0.382 |
0.566 |
| 78 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
-0.381 |
-0.025 |
| 79 |
257925_at |
AT3G23170
|
unknown |
-0.380 |
0.364 |
| 80 |
263837_at |
AT2G04500
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.376 |
0.081 |
| 81 |
249583_at |
AT5G37770
|
CML24, CALMODULIN-LIKE 24, TCH2, TOUCH 2 |
-0.374 |
0.145 |
| 82 |
250435_at |
AT5G10380
|
ATRING1, RING1 |
-0.368 |
0.370 |
| 83 |
255064_at |
AT4G08950
|
EXO, EXORDIUM |
-0.360 |
0.236 |
| 84 |
252557_at |
AT3G45960
|
ATEXLA3, expansin-like A3, ATEXPL3, ATHEXP BETA 2.3, EXLA3, expansin-like A3, EXPL3 |
-0.357 |
0.128 |
| 85 |
251192_at |
AT3G62720
|
ATXT1, XT1, xylosyltransferase 1, XXT1, XYG XYLOSYLTRANSFERASE 1 |
-0.350 |
0.199 |
| 86 |
265276_at |
AT2G28400
|
unknown |
-0.337 |
0.671 |
| 87 |
253915_at |
AT4G27280
|
[Calcium-binding EF-hand family protein] |
-0.337 |
0.489 |
| 88 |
255733_at |
AT1G25400
|
unknown |
-0.336 |
-0.049 |
| 89 |
255695_at |
AT4G00080
|
UNE11, unfertilized embryo sac 11 |
-0.336 |
-0.122 |