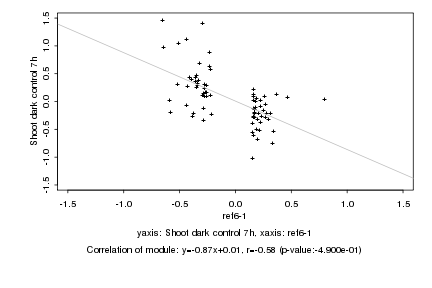
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ref6-1 |
Log2 signal ratio
Shoot dark control 7h |
| 1 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
0.790 |
0.050 |
| 2 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
0.462 |
0.082 |
| 3 |
251986_at |
AT3G53310
|
[AP2/B3-like transcriptional factor family protein] |
0.359 |
0.139 |
| 4 |
257644_at |
AT3G25780
|
AOC3, allene oxide cyclase 3 |
0.336 |
-0.522 |
| 5 |
261037_at |
AT1G17420
|
ATLOX3, Arabidopsis thaliana lipoxygenase 3, LOX3, lipoxygenase 3 |
0.326 |
-0.754 |
| 6 |
257919_at |
AT3G23250
|
ATMYB15, MYB DOMAIN PROTEIN 15, ATY19, MYB15, myb domain protein 15 |
0.310 |
-0.211 |
| 7 |
253643_at |
AT4G29780
|
unknown |
0.291 |
-0.313 |
| 8 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
0.276 |
-0.206 |
| 9 |
250098_at |
AT5G17350
|
unknown |
0.267 |
-0.271 |
| 10 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
0.266 |
-0.039 |
| 11 |
260973_at |
AT1G53490
|
HEI10, homolog of human HEI10 ( Enhancer of cell Invasion No.10) |
0.255 |
0.099 |
| 12 |
250292_at |
AT5G13220
|
JAS1, JASMONATE-ASSOCIATED 1, JAZ10, jasmonate-zim-domain protein 10, TIFY9, TIFY DOMAIN PROTEIN 9 |
0.250 |
-0.147 |
| 13 |
265723_at |
AT2G32140
|
[transmembrane receptors] |
0.231 |
-0.265 |
| 14 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
0.222 |
0.030 |
| 15 |
249245_at |
AT5G42280
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.222 |
-0.078 |
| 16 |
267411_at |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
0.218 |
-0.363 |
| 17 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
0.213 |
-0.519 |
| 18 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
0.205 |
-0.200 |
| 19 |
260399_at |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
0.197 |
-0.667 |
| 20 |
266316_at |
AT2G27080
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.197 |
-0.310 |
| 21 |
246949_at |
AT5G25140
|
CYP71B13, cytochrome P450, family 71, subfamily B, polypeptide 13 |
0.182 |
0.066 |
| 22 |
254926_at |
AT4G11280
|
ACS6, 1-aminocyclopropane-1-carboxylic acid (acc) synthase 6, ATACS6 |
0.182 |
-0.493 |
| 23 |
263236_at |
AT1G10470
|
ARR4, response regulator 4, ATRR1, RESPONCE REGULATOR 1, IBC7, INDUCED BY CYTOKININ 7, MEE7, maternal effect embryo arrest 7 |
0.173 |
-0.097 |
| 24 |
262743_at |
AT1G29020
|
[Calcium-binding EF-hand family protein] |
0.171 |
0.004 |
| 25 |
266246_at |
AT2G27690
|
CYP94C1, cytochrome P450, family 94, subfamily C, polypeptide 1 |
0.169 |
-0.277 |
| 26 |
249567_at |
AT5G38020
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.169 |
-0.141 |
| 27 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
0.169 |
-0.188 |
| 28 |
262211_at |
AT1G74930
|
ORA47 |
0.167 |
-0.205 |
| 29 |
254784_at |
AT4G12720
|
AtNUDT7, Arabidopsis thaliana Nudix hydrolase homolog 7, GFG1, GROWTH FACTOR GENE 1, NUDT7 |
0.161 |
0.034 |
| 30 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.160 |
0.092 |
| 31 |
250875_at |
AT5G04020
|
[calmodulin binding] |
0.155 |
0.225 |
| 32 |
253696_at |
AT4G29740
|
ATCKX4, CKX4, cytokinin oxidase 4 |
0.154 |
-0.253 |
| 33 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.154 |
-0.596 |
| 34 |
252679_at |
AT3G44260
|
AtCAF1a, CCR4- associated factor 1a, CAF1a, CCR4- associated factor 1a |
0.154 |
-0.113 |
| 35 |
253284_at |
AT4G34150
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.153 |
-0.281 |
| 36 |
264787_at |
AT2G17840
|
ERD7, EARLY-RESPONSIVE TO DEHYDRATION 7 |
0.153 |
0.130 |
| 37 |
267623_at |
AT2G39650
|
unknown |
0.152 |
-0.386 |
| 38 |
262383_at |
AT1G72940
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.150 |
-0.547 |
| 39 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
0.149 |
-1.011 |
| 40 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
-0.661 |
1.474 |
| 41 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
-0.646 |
0.987 |
| 42 |
264166_at |
AT1G65370
|
[TRAF-like family protein] |
-0.591 |
0.036 |
| 43 |
248888_at |
AT5G46240
|
AtKAT1, KAT1, potassium channel in Arabidopsis thaliana 1 |
-0.585 |
-0.190 |
| 44 |
247514_at |
AT5G61640
|
ATMSRA1, ARABIDOPSIS THALIANA METHIONINE SULFOXIDE REDUCTASE A1, PMSR1, peptidemethionine sulfoxide reductase 1 |
-0.522 |
0.309 |
| 45 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
-0.511 |
1.047 |
| 46 |
249427_at |
AT5G39850
|
[Ribosomal protein S4] |
-0.444 |
-0.064 |
| 47 |
250182_at |
AT5G14470
|
AtGALK2, GALK2, galactokinase 2 |
-0.441 |
1.121 |
| 48 |
251346_at |
AT3G60980
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.434 |
0.275 |
| 49 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
-0.414 |
0.432 |
| 50 |
252081_at |
AT3G51910
|
AT-HSFA7A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A7A, HSFA7A, heat shock transcription factor A7A |
-0.401 |
0.408 |
| 51 |
259561_at |
AT1G21250
|
AtWAK1, PRO25, WAK1, cell wall-associated kinase 1 |
-0.390 |
-0.267 |
| 52 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
-0.381 |
-0.210 |
| 53 |
255980_at |
AT1G33970
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.366 |
0.433 |
| 54 |
250748_at |
AT5G05710
|
[Pleckstrin homology (PH) domain superfamily protein] |
-0.362 |
0.364 |
| 55 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
-0.354 |
0.264 |
| 56 |
248208_at |
AT5G53980
|
ATHB52, homeobox protein 52, HB52, homeobox protein 52 |
-0.350 |
0.474 |
| 57 |
264902_at |
AT1G23060
|
MDP40, MICROTUBULE DESTABILIZING PROTEIN 40 |
-0.344 |
0.333 |
| 58 |
260771_at |
AT1G49160
|
WNK7 |
-0.342 |
0.299 |
| 59 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.336 |
0.393 |
| 60 |
248371_at |
AT5G51810
|
AT2353, ATGA20OX2, GA20OX2, gibberellin 20 oxidase 2 |
-0.330 |
0.684 |
| 61 |
248916_at |
AT5G45840
|
[Leucine-rich repeat protein kinase family protein] |
-0.301 |
0.111 |
| 62 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
-0.300 |
1.417 |
| 63 |
254251_at |
AT4G23300
|
CRK22, cysteine-rich RLK (RECEPTOR-like protein kinase) 22 |
-0.287 |
-0.121 |
| 64 |
257369_at |
AT2G35550
|
ATBPC7, BASIC PENTACYSTEINE 7, BBR, BPC7, basic pentacysteine 7 |
-0.287 |
0.159 |
| 65 |
259743_at |
AT1G71140
|
[MATE efflux family protein] |
-0.286 |
-0.340 |
| 66 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
-0.285 |
0.098 |
| 67 |
251623_at |
AT3G57390
|
AGL18, AGAMOUS-like 18 |
-0.279 |
0.239 |
| 68 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.277 |
0.310 |
| 69 |
247345_at |
AT5G63760
|
ARI15, ARIADNE 15, ATARI15, ARABIDOPSIS ARIADNE 15 |
-0.273 |
0.176 |
| 70 |
248236_at |
AT5G53870
|
AtENODL1, ENODL1, early nodulin-like protein 1 |
-0.266 |
0.301 |
| 71 |
260201_at |
AT1G67600
|
[Acid phosphatase/vanadium-dependent haloperoxidase-related protein] |
-0.263 |
0.180 |
| 72 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
-0.262 |
0.096 |
| 73 |
254197_at |
AT4G24040
|
ATTRE1, TRE1, trehalase 1 |
-0.235 |
0.883 |
| 74 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
-0.235 |
0.641 |
| 75 |
266156_at |
AT2G28110
|
FRA8, FRAGILE FIBER 8, IRX7, IRREGULAR XYLEM 7 |
-0.229 |
0.582 |
| 76 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
-0.229 |
0.116 |
| 77 |
251750_at |
AT3G55710
|
[UDP-Glycosyltransferase superfamily protein] |
-0.221 |
-0.231 |