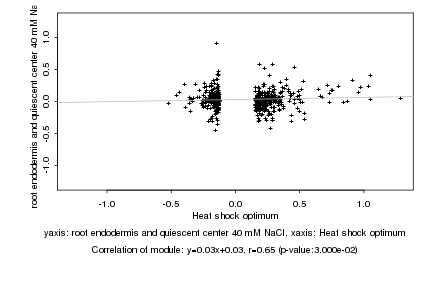
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Heat shock optimum |
Log2 signal ratio
root endodermis and quiescent center 40 mM NaCl |
| 1 |
266841_at |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
1.282 |
0.054 |
| 2 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
1.048 |
0.411 |
| 3 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
1.047 |
0.042 |
| 4 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
1.030 |
0.241 |
| 5 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
0.972 |
0.234 |
| 6 |
254839_at |
AT4G12400
|
Hop3, Hop3 |
0.955 |
0.156 |
| 7 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
0.911 |
0.337 |
| 8 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
0.868 |
0.010 |
| 9 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.841 |
-0.002 |
| 10 |
266590_at |
AT2G46240
|
ATBAG6, ARABIDOPSIS THALIANA BCL-2-ASSOCIATED ATHANOGENE 6, BAG6, BCL-2-associated athanogene 6 |
0.800 |
0.244 |
| 11 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
0.748 |
0.175 |
| 12 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
0.743 |
0.182 |
| 13 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
0.732 |
0.136 |
| 14 |
252081_at |
AT3G51910
|
AT-HSFA7A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A7A, HSFA7A, heat shock transcription factor A7A |
0.732 |
0.000 |
| 15 |
262307_at |
AT1G71000
|
[Chaperone DnaJ-domain superfamily protein] |
0.711 |
0.262 |
| 16 |
254878_at |
AT4G11660
|
AT-HSFB2B, HSF7, HSFB2B, HEAT SHOCK TRANSCRIPTION FACTOR B2B |
0.675 |
0.075 |
| 17 |
256518_at |
AT1G66080
|
unknown |
0.662 |
0.084 |
| 18 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
0.662 |
0.087 |
| 19 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
0.640 |
0.199 |
| 20 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
0.531 |
-0.266 |
| 21 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
0.531 |
-0.179 |
| 22 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.528 |
0.318 |
| 23 |
260881_at |
AT1G21550
|
[Calcium-binding EF-hand family protein] |
0.519 |
-0.011 |
| 24 |
259037_at |
AT3G09350
|
Fes1A, Fes1A |
0.509 |
0.201 |
| 25 |
258336_at |
AT3G16050
|
A37, ATPDX1.2, ARABIDOPSIS THALIANA PYRIDOXINE BIOSYNTHESIS 1.2, PDX1.2, pyridoxine biosynthesis 1.2 |
0.502 |
-0.015 |
| 26 |
261684_at |
AT1G47400
|
unknown |
0.496 |
0.035 |
| 27 |
260399_at |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
0.493 |
-0.147 |
| 28 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
0.491 |
0.037 |
| 29 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.491 |
0.097 |
| 30 |
261265_at |
AT1G26800
|
[RING/U-box superfamily protein] |
0.484 |
0.168 |
| 31 |
261081_at |
AT1G07350
|
SR45a, serine/arginine rich-like protein 45a |
0.483 |
0.090 |
| 32 |
266415_at |
AT2G38530
|
cdf3, cell growth defect factor-3, LP2, LTP2, lipid transfer protein 2 |
0.482 |
-0.099 |
| 33 |
262656_at |
AT1G14200
|
[RING/U-box superfamily protein] |
0.481 |
0.061 |
| 34 |
256526_at |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
0.457 |
0.036 |
| 35 |
257644_at |
AT3G25780
|
AOC3, allene oxide cyclase 3 |
0.457 |
-0.021 |
| 36 |
249575_at |
AT5G37670
|
[HSP20-like chaperones superfamily protein] |
0.454 |
0.545 |
| 37 |
264019_at |
AT2G21130
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
0.448 |
0.137 |
| 38 |
254233_at |
AT4G23800
|
3xHMG-box2, 3xHigh Mobility Group-box2 |
0.433 |
-0.307 |
| 39 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
0.433 |
0.099 |
| 40 |
262978_at |
AT1G75780
|
TUB1, tubulin beta-1 chain |
0.429 |
-0.204 |
| 41 |
246018_at |
AT5G10695
|
unknown |
0.427 |
0.062 |
| 42 |
259479_at |
AT1G19020
|
unknown |
0.425 |
0.134 |
| 43 |
260025_at |
AT1G30070
|
[SGS domain-containing protein] |
0.422 |
0.102 |
| 44 |
257154_at |
AT3G27210
|
unknown |
0.417 |
0.006 |
| 45 |
256999_at |
AT3G14200
|
[Chaperone DnaJ-domain superfamily protein] |
0.411 |
0.164 |
| 46 |
258939_at |
AT3G10020
|
unknown |
0.407 |
0.195 |
| 47 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
0.394 |
0.254 |
| 48 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
0.390 |
0.350 |
| 49 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
0.377 |
0.208 |
| 50 |
254914_at |
AT4G11290
|
[Peroxidase superfamily protein] |
0.374 |
-0.065 |
| 51 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
0.370 |
-0.105 |
| 52 |
259272_at |
AT3G01290
|
AtHIR2, HIR2, hypersensitive induced reaction 2 |
0.363 |
0.081 |
| 53 |
264787_at |
AT2G17840
|
ERD7, EARLY-RESPONSIVE TO DEHYDRATION 7 |
0.362 |
0.159 |
| 54 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
0.360 |
0.229 |
| 55 |
250292_at |
AT5G13220
|
JAS1, JASMONATE-ASSOCIATED 1, JAZ10, jasmonate-zim-domain protein 10, TIFY9, TIFY DOMAIN PROTEIN 9 |
0.358 |
-0.042 |
| 56 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
0.356 |
-0.000 |
| 57 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
0.350 |
0.143 |
| 58 |
246944_at |
AT5G25450
|
[Cytochrome bd ubiquinol oxidase, 14kDa subunit] |
0.350 |
0.309 |
| 59 |
261037_at |
AT1G17420
|
ATLOX3, Arabidopsis thaliana lipoxygenase 3, LOX3, lipoxygenase 3 |
0.348 |
0.039 |
| 60 |
246612_at |
AT5G35320
|
unknown |
0.346 |
0.141 |
| 61 |
255607_at |
AT4G01130
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.341 |
0.015 |
| 62 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
0.341 |
-0.123 |
| 63 |
247425_at |
AT5G62550
|
unknown |
0.341 |
-0.134 |
| 64 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
0.339 |
0.020 |
| 65 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
0.336 |
0.076 |
| 66 |
263498_at |
AT2G42610
|
LSH10, LIGHT SENSITIVE HYPOCOTYLS 10 |
0.334 |
0.075 |
| 67 |
259609_at |
AT1G52410
|
AtTSA1, TSA1, TSK-associating protein 1 |
0.331 |
-0.053 |
| 68 |
267411_at |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
0.328 |
-0.022 |
| 69 |
251432_at |
AT3G59820
|
AtLETM1, LETM1, leucine zipper-EF-hand-containing transmembrane protein 1 |
0.324 |
0.049 |
| 70 |
254255_at |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
0.318 |
0.015 |
| 71 |
251494_at |
AT3G59350
|
[Protein kinase superfamily protein] |
0.313 |
0.060 |
| 72 |
265442_at |
AT2G20940
|
unknown |
0.310 |
-0.009 |
| 73 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
0.310 |
-0.008 |
| 74 |
245602_at |
AT4G14270
|
unknown |
0.307 |
0.103 |
| 75 |
250036_at |
AT5G18340
|
[ARM repeat superfamily protein] |
0.303 |
0.095 |
| 76 |
253636_at |
AT4G30500
|
unknown |
0.302 |
0.025 |
| 77 |
253666_at |
AT4G30270
|
MERI-5, MERISTEM 5, MERI5B, meristem-5, SEN4, SENESCENCE 4, XTH24, xyloglucan endotransglucosylase/hydrolase 24 |
0.300 |
0.069 |
| 78 |
247137_at |
AT5G66210
|
CPK28, calcium-dependent protein kinase 28 |
0.300 |
-0.082 |
| 79 |
249719_at |
AT5G35735
|
[Auxin-responsive family protein] |
0.299 |
0.106 |
| 80 |
245076_at |
AT2G23170
|
GH3.3 |
0.299 |
0.203 |
| 81 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
0.298 |
0.011 |
| 82 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
0.298 |
0.045 |
| 83 |
245688_at |
AT1G28290
|
AGP31, arabinogalactan protein 31 |
0.297 |
-0.141 |
| 84 |
245749_at |
AT1G51090
|
[Heavy metal transport/detoxification superfamily protein ] |
0.296 |
0.243 |
| 85 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
0.296 |
0.151 |
| 86 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
0.294 |
-0.007 |
| 87 |
259327_at |
AT3G16460
|
JAL34, jacalin-related lectin 34 |
0.292 |
-0.092 |
| 88 |
257822_at |
AT3G25230
|
ATFKBP62, FKBP62, FK506 BINDING PROTEIN 62, ROF1, rotamase FKBP 1 |
0.291 |
0.049 |
| 89 |
261167_at |
AT1G04980
|
ATPDI10, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 10, ATPDIL2-2, PDI-like 2-2, PDI10, PROTEIN DISULFIDE ISOMERASE, PDIL2-2, PDI-like 2-2 |
0.290 |
-0.143 |
| 90 |
258794_at |
AT3G04710
|
TPR10, tetratricopeptide repeat 10 |
0.290 |
0.050 |
| 91 |
262548_at |
AT1G31280
|
AGO2, argonaute 2, AtAGO2 |
0.287 |
0.094 |
| 92 |
247189_at |
AT5G65390
|
AGP7, arabinogalactan protein 7 |
0.286 |
-0.287 |
| 93 |
250228_at |
AT5G13840
|
CCS52B, cell cycle switch protein 52 B, FZR3, FIZZY-related 3 |
0.286 |
-0.262 |
| 94 |
256114_at |
AT1G16850
|
unknown |
0.284 |
0.588 |
| 95 |
250823_at |
AT5G05180
|
unknown |
0.282 |
-0.292 |
| 96 |
249456_at |
AT5G39410
|
[Saccharopine dehydrogenase ] |
0.282 |
0.049 |
| 97 |
267336_at |
AT2G19310
|
[HSP20-like chaperones superfamily protein] |
0.282 |
0.244 |
| 98 |
250279_at |
AT5G13200
|
[GRAM domain family protein] |
0.281 |
0.163 |
| 99 |
254211_at |
AT4G23570
|
SGT1A |
0.279 |
0.040 |
| 100 |
252114_at |
AT3G51450
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.279 |
-0.194 |
| 101 |
252053_at |
AT3G52400
|
ATSYP122, SYP122, syntaxin of plants 122 |
0.277 |
-0.003 |
| 102 |
252234_at |
AT3G49780
|
ATPSK3 (FORMER SYMBOL), ATPSK4, phytosulfokine 4 precursor, PSK4, phytosulfokine 4 precursor |
0.276 |
0.112 |
| 103 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
0.275 |
-0.003 |
| 104 |
245789_at |
AT1G32090
|
[early-responsive to dehydration stress protein (ERD4)] |
0.273 |
-0.078 |
| 105 |
248197_at |
AT5G54190
|
PORA, protochlorophyllide oxidoreductase A |
0.271 |
-0.009 |
| 106 |
264045_at |
AT2G22450
|
AtRIBA2, RIBA2, homolog of ribA 2 |
0.270 |
0.028 |
| 107 |
247348_at |
AT5G63810
|
AtBGAL10, Arabidopsis thaliana beta-galactosidase 10, BGAL10, beta-galactosidase 10 |
0.269 |
-0.032 |
| 108 |
245343_at |
AT4G15830
|
[ARM repeat superfamily protein] |
0.266 |
-0.408 |
| 109 |
250412_at |
AT5G11150
|
ATVAMP713, vesicle-associated membrane protein 713, VAMP713, VAMP713, vesicle-associated membrane protein 713 |
0.266 |
0.069 |
| 110 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
0.264 |
0.407 |
| 111 |
266613_at |
AT2G14900
|
[Gibberellin-regulated family protein] |
0.264 |
-0.027 |
| 112 |
247280_at |
AT5G64260
|
EXL2, EXORDIUM like 2 |
0.261 |
0.074 |
| 113 |
266261_at |
AT2G27580
|
[A20/AN1-like zinc finger family protein] |
0.261 |
0.067 |
| 114 |
246415_at |
AT5G17160
|
unknown |
0.261 |
-0.198 |
| 115 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
0.261 |
0.162 |
| 116 |
246550_at |
AT5G14920
|
GASA14, A-stimulated in Arabidopsis 14 |
0.259 |
-0.174 |
| 117 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.258 |
0.210 |
| 118 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
0.257 |
-0.152 |
| 119 |
253687_at |
AT4G29520
|
[LOCATED IN: endoplasmic reticulum, plasma membrane] |
0.256 |
-0.215 |
| 120 |
255236_at |
AT4G05520
|
ATEHD2, EPS15 homology domain 2, EHD2, EPS15 homology domain 2 |
0.254 |
0.045 |
| 121 |
264968_at |
AT1G67360
|
[Rubber elongation factor protein (REF)] |
0.252 |
0.139 |
| 122 |
258830_at |
AT3G07090
|
[PPPDE putative thiol peptidase family protein] |
0.252 |
0.101 |
| 123 |
261564_at |
AT1G01720
|
ANAC002, Arabidopsis NAC domain containing protein 2, ATAF1 |
0.251 |
0.013 |
| 124 |
258923_at |
AT3G10450
|
SCPL7, serine carboxypeptidase-like 7 |
0.249 |
0.034 |
| 125 |
263548_at |
AT2G21660
|
ATGRP7, GLYCINE RICH PROTEIN 7, CCR2, cold, circadian rhythm, and rna binding 2, GR-RBP7, GLYCINE-RICH RNA-BINDING PROTEIN 7, GRP7, GLYCINE-RICH RNA-BINDING PROTEIN 7 |
0.248 |
-0.048 |
| 126 |
251975_at |
AT3G53230
|
AtCDC48B, cell division cycle 48B |
0.243 |
0.143 |
| 127 |
260656_at |
AT1G19380
|
unknown |
0.243 |
0.026 |
| 128 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
0.243 |
0.140 |
| 129 |
262159_at |
AT1G52720
|
unknown |
0.243 |
0.041 |
| 130 |
264402_at |
AT2G25140
|
CLPB-M, CASEIN LYTIC PROTEINASE B-M, CLPB4, casein lytic proteinase B4, HSP98.7, HEAT SHOCK PROTEIN 98.7 |
0.241 |
0.060 |
| 131 |
266977_at |
AT2G39420
|
[alpha/beta-Hydrolases superfamily protein] |
0.241 |
-0.016 |
| 132 |
261780_at |
AT1G76310
|
CYCB2;4, CYCLIN B2;4 |
0.240 |
-0.291 |
| 133 |
252684_at |
AT3G44400
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.237 |
-0.120 |
| 134 |
261086_at |
AT1G17460
|
TRFL3, TRF-like 3 |
0.237 |
-0.058 |
| 135 |
264217_at |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
0.237 |
0.058 |
| 136 |
264663_at |
AT1G09970
|
LRR XI-23, RLK7, receptor-like kinase 7 |
0.236 |
0.045 |
| 137 |
261368_at |
AT1G53070
|
[Legume lectin family protein] |
0.235 |
0.025 |
| 138 |
262014_at |
AT1G35660
|
unknown |
0.235 |
0.089 |
| 139 |
255410_at |
AT4G03100
|
[Rho GTPase activating protein with PAK-box/P21-Rho-binding domain] |
0.235 |
-0.031 |
| 140 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
0.234 |
-0.125 |
| 141 |
261125_at |
AT1G04990
|
[Zinc finger C-x8-C-x5-C-x3-H type family protein] |
0.234 |
0.126 |
| 142 |
265359_at |
AT2G16720
|
ATMYB7, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 7, ATY49, MYB7, myb domain protein 7 |
0.232 |
-0.013 |
| 143 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.232 |
0.101 |
| 144 |
258225_at |
AT3G15630
|
unknown |
0.232 |
0.036 |
| 145 |
254964_at |
AT4G11080
|
3xHMG-box1, 3xHigh Mobility Group-box1 |
0.231 |
-0.265 |
| 146 |
259723_at |
AT1G60960
|
ATIRT3, IRON REGULATED TRANSPORTER 3, IRT3, iron regulated transporter 3 |
0.230 |
-0.030 |
| 147 |
246779_at |
AT5G27520
|
AtPNC2, PNC2, peroxisomal adenine nucleotide carrier 2 |
0.229 |
0.051 |
| 148 |
266009_at |
AT2G37420
|
[ATP binding microtubule motor family protein] |
0.228 |
-0.034 |
| 149 |
263513_at |
AT2G12400
|
unknown |
0.228 |
-0.125 |
| 150 |
248698_at |
AT5G48380
|
BIR1, BAK1-interacting receptor-like kinase 1 |
0.227 |
0.095 |
| 151 |
258262_at |
AT3G15770
|
unknown |
0.227 |
0.077 |
| 152 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
0.225 |
0.518 |
| 153 |
267036_at |
AT2G38465
|
unknown |
0.225 |
0.077 |
| 154 |
248045_at |
AT5G56030
|
AtHsp90.2, HEAT SHOCK PROTEIN 90.2, ERD8, EARLY-RESPONSIVE TO DEHYDRATION 8, HSP81-2, heat shock protein 81-2, HSP81.2, heat shock protein 81.2, HSP90.2, HEAT SHOCK PROTEIN 90.2 |
0.224 |
-0.038 |
| 155 |
265530_at |
AT2G06050
|
AtOPR3, DDE1, DELAYED DEHISCENCE 1, OPR3, oxophytodienoate-reductase 3 |
0.223 |
-0.083 |
| 156 |
264465_at |
AT1G10230
|
ASK18, SKP1-like 18, SK18, SKP1-like 18 |
0.222 |
-0.067 |
| 157 |
267483_at |
AT2G02810
|
ATUTR1, UDP-GALACTOSE TRANSPORTER 1, UTR1, UDP-galactose transporter 1 |
0.221 |
-0.150 |
| 158 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.221 |
0.278 |
| 159 |
267083_at |
AT2G41100
|
ATCAL4, ARABIDOPSIS THALIANA CALMODULIN LIKE 4, TCH3, TOUCH 3 |
0.220 |
-0.008 |
| 160 |
259137_at |
AT3G10300
|
[Calcium-binding EF-hand family protein] |
0.220 |
0.037 |
| 161 |
245244_at |
AT1G44350
|
ILL6, IAA-leucine resistant (ILR)-like gene 6 |
0.219 |
0.148 |
| 162 |
266839_at |
AT2G25930
|
ELF3, EARLY FLOWERING 3, PYK20 |
0.219 |
0.033 |
| 163 |
258915_at |
AT3G10640
|
VPS60.1 |
0.219 |
-0.019 |
| 164 |
263120_at |
AT1G78490
|
CYP708A3, cytochrome P450, family 708, subfamily A, polypeptide 3 |
0.219 |
0.105 |
| 165 |
245686_at |
AT5G22060
|
ATJ2, ARABIDOPSIS THALIANA DNAJ HOMOLOGUE 2, J2, DNAJ homologue 2 |
0.219 |
0.025 |
| 166 |
266119_at |
AT2G02100
|
LCR69, low-molecular-weight cysteine-rich 69, PDF2.2 |
0.218 |
-0.055 |
| 167 |
261272_at |
AT1G26665
|
[Mediator complex, subunit Med10] |
0.217 |
0.027 |
| 168 |
249837_at |
AT5G23480
|
[SWIB/MDM2 domain] |
0.216 |
0.011 |
| 169 |
248410_at |
AT5G51570
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
0.216 |
0.022 |
| 170 |
263535_at |
AT2G24970
|
unknown |
0.216 |
-0.014 |
| 171 |
247279_at |
AT5G64310
|
AGP1, arabinogalactan protein 1, ATAGP1 |
0.216 |
0.029 |
| 172 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.214 |
0.273 |
| 173 |
254377_at |
AT4G21650
|
[Subtilase family protein] |
0.213 |
-0.011 |
| 174 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
0.213 |
-0.106 |
| 175 |
245247_at |
AT4G17230
|
SCL13, SCARECROW-like 13 |
0.213 |
-0.013 |
| 176 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
0.213 |
0.238 |
| 177 |
259640_at |
AT1G52400
|
ATBG1, A. THALIANA BETA-GLUCOSIDASE 1, BGL1, BETA-GLUCOSIDASE HOMOLOG 1, BGLU18, beta glucosidase 18 |
0.210 |
0.014 |
| 178 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
0.210 |
0.051 |
| 179 |
248258_at |
AT5G53400
|
BOB1, BOBBER1 |
0.209 |
0.007 |
| 180 |
263800_at |
AT2G24600
|
[Ankyrin repeat family protein] |
0.209 |
-0.188 |
| 181 |
257748_at |
AT3G18710
|
ATPUB29, ARABIDOPSIS THALIANA PLANT U-BOX 29, PUB29, plant U-box 29 |
0.208 |
-0.119 |
| 182 |
253351_at |
AT4G33700
|
unknown |
0.208 |
0.059 |
| 183 |
261485_at |
AT1G14360
|
ATUTR3, UTR3, UDP-galactose transporter 3 |
0.208 |
-0.119 |
| 184 |
259169_at |
AT3G03520
|
NPC3, non-specific phospholipase C3 |
0.207 |
0.139 |
| 185 |
255485_at |
AT4G02550
|
unknown |
0.206 |
0.031 |
| 186 |
266299_at |
AT2G29450
|
AT103-1A, ATGSTU1, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 1, ATGSTU5, glutathione S-transferase tau 5, GSTU5, glutathione S-transferase tau 5 |
0.205 |
0.150 |
| 187 |
250931_at |
AT5G03200
|
LUL1, LOG2-LIKE UBIQUITIN LIGASE1 |
0.205 |
-0.080 |
| 188 |
267280_at |
AT2G19450
|
ABX45, AS11, ATDGAT, Arabidopsis thaliana acyl-CoA:diacylglycerol acyltransferase, AtDGAT1, DGAT1, acyl-CoA:diacylglycerol acyltransferase 1, RDS1, TAG1, TRIACYLGLYCEROL BIOSYNTHESIS DEFECT 1 |
0.205 |
0.100 |
| 189 |
253630_at |
AT4G30490
|
[AFG1-like ATPase family protein] |
0.204 |
0.016 |
| 190 |
247786_at |
AT5G58600
|
PMR5, POWDERY MILDEW RESISTANT 5, TBL44, TRICHOME BIREFRINGENCE-LIKE 44 |
0.203 |
0.015 |
| 191 |
266423_at |
AT2G41340
|
RPB5D, RNA polymerase II fifth largest subunit, D |
0.202 |
0.043 |
| 192 |
252213_at |
AT3G50210
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.200 |
0.006 |
| 193 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
0.199 |
0.012 |
| 194 |
246858_at |
AT5G25930
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.199 |
0.023 |
| 195 |
254784_at |
AT4G12720
|
AtNUDT7, Arabidopsis thaliana Nudix hydrolase homolog 7, GFG1, GROWTH FACTOR GENE 1, NUDT7 |
0.198 |
-0.044 |
| 196 |
245272_at |
AT4G17250
|
unknown |
0.198 |
-0.026 |
| 197 |
262671_at |
AT1G76040
|
CPK29, calcium-dependent protein kinase 29 |
0.198 |
-0.206 |
| 198 |
265077_at |
AT1G55530
|
[RING/U-box superfamily protein] |
0.195 |
0.114 |
| 199 |
253403_at |
AT4G32830
|
AtAUR1, ataurora1, AUR1, ataurora1 |
0.195 |
-0.125 |
| 200 |
255232_at |
AT4G05330
|
AGD13, ARF-GAP domain 13 |
0.194 |
-0.127 |
| 201 |
267141_at |
AT2G38090
|
[Duplicated homeodomain-like superfamily protein] |
0.193 |
-0.037 |
| 202 |
247187_at |
AT5G65490
|
unknown |
0.193 |
0.136 |
| 203 |
260395_at |
AT1G69780
|
ATHB13 |
0.191 |
-0.083 |
| 204 |
250661_at |
AT5G07030
|
[Eukaryotic aspartyl protease family protein] |
0.191 |
-0.100 |
| 205 |
256663_at |
AT3G12050
|
[Aha1 domain-containing protein] |
0.190 |
0.001 |
| 206 |
248793_at |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
0.190 |
0.056 |
| 207 |
251563_at |
AT3G57880
|
[Calcium-dependent lipid-binding (CaLB domain) plant phosphoribosyltransferase family protein] |
0.189 |
0.030 |
| 208 |
256743_at |
AT3G29370
|
P1R3, P1R3 |
0.189 |
-0.060 |
| 209 |
265999_at |
AT2G24100
|
ASG1, ALTERED SEED GERMINATION 1 |
0.188 |
0.141 |
| 210 |
247910_at |
AT5G57410
|
[Afadin/alpha-actinin-binding protein] |
0.188 |
0.103 |
| 211 |
252149_at |
AT3G51290
|
APSR1, Altered Phosphate Starvation Response 1 |
0.186 |
-0.138 |
| 212 |
247937_at |
AT5G57110
|
ACA8, autoinhibited Ca2+ -ATPase, isoform 8, AT-ACA8, AUTOINHIBITED CA2+ -ATPASE, ISOFORM 8 |
0.186 |
0.159 |
| 213 |
266959_at |
AT2G07690
|
MCM5, MINICHROMOSOME MAINTENANCE 5 |
0.186 |
-0.043 |
| 214 |
251584_at |
AT3G58620
|
TTL4, tetratricopetide-repeat thioredoxin-like 4 |
0.186 |
-0.296 |
| 215 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
0.185 |
0.031 |
| 216 |
265510_at |
AT2G05630
|
ATG8D |
0.185 |
0.051 |
| 217 |
247056_at |
AT5G66750
|
ATDDM1, CHA1, CHR01, CHROMATIN REMODELING 1, CHR1, chromatin remodeling 1, DDM1, DECREASED DNA METHYLATION 1, SOM1, SOMNIFEROUS 1, SOM4 |
0.185 |
0.046 |
| 218 |
262756_at |
AT1G16370
|
ATOCT6, ARABIDOPSIS THALIANA ORGANIC CATION/CARNITINE TRANSPORTER 6, OCT6, organic cation/carnitine transporter 6 |
0.184 |
-0.110 |
| 219 |
267073_at |
AT2G41160
|
[Ubiquitin-associated (UBA) protein] |
0.184 |
0.056 |
| 220 |
253331_at |
AT4G33490
|
[Eukaryotic aspartyl protease family protein] |
0.184 |
0.008 |
| 221 |
262892_at |
AT1G79440
|
ALDH5F1, aldehyde dehydrogenase 5F1, ENF1, ENLARGED FIL EXPRESSION DOMAIN 1, SSADH, SUCCINIC SEMIALDEHYDE DEHYDROGENASE, SSADH1, SUCCINIC SEMIALDEHYDE DEHYDROGENASE 1 |
0.183 |
-0.075 |
| 222 |
253981_at |
AT4G26670
|
[Mitochondrial import inner membrane translocase subunit Tim17/Tim22/Tim23 family protein] |
0.183 |
0.062 |
| 223 |
250502_at |
AT5G09590
|
HSC70-5, HEAT SHOCK COGNATE, MTHSC70-2, mitochondrial HSO70 2 |
0.182 |
0.041 |
| 224 |
259978_at |
AT1G76540
|
CDKB2;1, cyclin-dependent kinase B2;1 |
0.182 |
-0.094 |
| 225 |
264624_at |
AT1G08930
|
ERD6, EARLY RESPONSE TO DEHYDRATION 6 |
0.182 |
0.009 |
| 226 |
263515_at |
AT2G21640
|
unknown |
0.181 |
0.074 |
| 227 |
250828_at |
AT5G05250
|
unknown |
0.181 |
-0.184 |
| 228 |
246288_at |
AT1G31850
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.181 |
0.014 |
| 229 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
0.181 |
0.584 |
| 230 |
247962_at |
AT5G56580
|
ANQ1, ARABIDOPSIS NQK1, ATMKK6, ARABIDOPSIS THALIANA MAP KINASE KINASE 6, MKK6, MAP kinase kinase 6 |
0.180 |
0.003 |
| 231 |
265675_at |
AT2G32120
|
HSP70T-2, heat-shock protein 70T-2 |
0.180 |
0.217 |
| 232 |
258063_at |
AT3G14620
|
CYP72A8, cytochrome P450, family 72, subfamily A, polypeptide 8 |
0.180 |
0.076 |
| 233 |
258406_at |
AT3G17611
|
ATRBL10, RHOMBOID-like protein 10, ATRBL14, RHOMBOID-like protein 14, RBL10, RHOMBOID-like protein 10, RBL14, RHOMBOID-like protein 14 |
0.180 |
0.097 |
| 234 |
250565_at |
AT5G08000
|
E13L3, glucan endo-1,3-beta-glucosidase-like protein 3, PDCB2, PLASMODESMATA CALLOSE-BINDING PROTEIN 2 |
0.179 |
0.106 |
| 235 |
257715_at |
AT3G12750
|
AtZIP1, ZIP1, zinc transporter 1 precursor |
0.179 |
0.224 |
| 236 |
258979_at |
AT3G09440
|
[Heat shock protein 70 (Hsp 70) family protein] |
0.178 |
0.118 |
| 237 |
260943_at |
AT1G45145
|
ATH5, THIOREDOXIN H-TYPE 5, ATTRX5, thioredoxin H-type 5, LIV1, LOCUS OF INSENSITIVITY TO VICTORIN 1, TRX-h5, THIOREDOXIN H-TYPE 5, TRX5, thioredoxin H-type 5 |
0.177 |
0.107 |
| 238 |
250424_at |
AT5G10550
|
GTE2, global transcription factor group E2 |
0.177 |
-0.044 |
| 239 |
266456_at |
AT2G22770
|
NAI1 |
0.177 |
-0.254 |
| 240 |
267264_at |
AT2G22970
|
SCPL11, serine carboxypeptidase-like 11 |
0.177 |
-0.312 |
| 241 |
246002_at |
AT5G20740
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.177 |
0.017 |
| 242 |
246253_at |
AT4G37260
|
ATMYB73, MYB73, myb domain protein 73 |
0.176 |
-0.022 |
| 243 |
245828_at |
AT1G57820
|
ORTH2, ORTHRUS 2, VIM1, VARIANT IN METHYLATION 1 |
0.176 |
-0.004 |
| 244 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
0.175 |
0.126 |
| 245 |
250127_at |
AT5G16380
|
unknown |
0.175 |
0.081 |
| 246 |
246929_at |
AT5G25210
|
unknown |
0.175 |
0.059 |
| 247 |
247043_at |
AT5G66880
|
SNRK2-3, SUCROSE NONFERMENTING 1 (SNF1)-RELATED PROTEIN KINASE 2-3, SNRK2.3, sucrose nonfermenting 1(SNF1)-related protein kinase 2.3, SRK2I |
0.175 |
0.106 |
| 248 |
245821_at |
AT1G26270
|
[Phosphatidylinositol 3- and 4-kinase family protein] |
0.175 |
0.051 |
| 249 |
260472_at |
AT1G10990
|
unknown |
0.175 |
0.018 |
| 250 |
259911_at |
AT1G72680
|
ATCAD1, CINNAMYL ALCOHOL DEHYDROGENASE 1, CAD1, cinnamyl-alcohol dehydrogenase |
0.175 |
0.167 |
| 251 |
262312_at |
AT1G70830
|
MLP28, MLP-like protein 28 |
0.175 |
0.013 |
| 252 |
260728_at |
AT1G48210
|
[Protein kinase superfamily protein] |
0.174 |
0.035 |
| 253 |
267462_at |
AT2G33735
|
[Chaperone DnaJ-domain superfamily protein] |
0.174 |
0.014 |
| 254 |
248774_at |
AT5G47830
|
unknown |
0.172 |
0.060 |
| 255 |
259383_at |
AT3G16470
|
JAL35, jacalin-related lectin 35, JR1, JASMONATE RESPONSIVE 1 |
0.172 |
-0.082 |
| 256 |
253949_at |
AT4G26780
|
AR192, MGE2, mitochondrial GrpE 2 |
0.172 |
-0.090 |
| 257 |
248021_at |
AT5G56500
|
Cpn60beta3, chaperonin-60beta3 |
0.171 |
-0.027 |
| 258 |
267355_at |
AT2G39900
|
WLIM2a, WLIM2a |
0.170 |
-0.110 |
| 259 |
249746_at |
AT5G24590
|
ANAC091, Arabidopsis NAC domain containing protein 91, TIP, TCV-interacting protein |
0.169 |
-0.014 |
| 260 |
264246_at |
AT1G60140
|
ATTPS10, trehalose phosphate synthase, TPS10, trehalose phosphate synthase, TPS10, TREHALOSE PHOSPHATE SYNTHASE 10 |
0.169 |
0.002 |
| 261 |
260462_at |
AT1G10970
|
ATZIP4, ZIP4, zinc transporter 4 precursor |
0.169 |
0.092 |
| 262 |
249372_at |
AT5G40760
|
G6PD6, glucose-6-phosphate dehydrogenase 6 |
0.169 |
-0.002 |
| 263 |
247835_at |
AT5G57910
|
unknown |
0.168 |
0.099 |
| 264 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
0.168 |
-0.213 |
| 265 |
262643_at |
AT1G62770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.168 |
-0.035 |
| 266 |
250580_at |
AT5G07440
|
GDH2, glutamate dehydrogenase 2 |
0.167 |
0.012 |
| 267 |
256433_at |
AT3G10985
|
ATWI-12, ARABIDOPSIS THALIANA WOUND-INDUCED PROTEIN 12, SAG20, senescence associated gene 20, WI12, WOUND-INDUCED PROTEIN 12 |
0.167 |
0.015 |
| 268 |
254190_at |
AT4G23885
|
unknown |
0.167 |
-0.055 |
| 269 |
259570_at |
AT1G20440
|
AtCOR47, COR47, cold-regulated 47, RD17 |
0.166 |
0.104 |
| 270 |
261713_at |
AT1G32640
|
ATMYC2, JAI1, JASMONATE INSENSITIVE 1, JIN1, JASMONATE INSENSITIVE 1, MYC2, RD22BP1, ZBF1 |
0.166 |
-0.075 |
| 271 |
255255_at |
AT4G05070
|
[Wound-responsive family protein] |
0.165 |
0.067 |
| 272 |
251789_at |
AT3G55450
|
PBL1, PBS1-like 1 |
0.164 |
0.098 |
| 273 |
248138_at |
AT5G54960
|
PDC2, pyruvate decarboxylase-2 |
0.164 |
0.106 |
| 274 |
260546_at |
AT2G43520
|
ATTI2, trypsin inhibitor protein 2, TI2, trypsin inhibitor protein 2 |
0.164 |
0.090 |
| 275 |
262802_at |
AT1G20930
|
CDKB2;2, cyclin-dependent kinase B2;2 |
0.163 |
-0.060 |
| 276 |
249293_at |
AT5G41260
|
BSK8, brassinosteroid-signaling kinase 8 |
0.163 |
-0.148 |
| 277 |
252464_at |
AT3G47160
|
[RING/U-box superfamily protein] |
0.163 |
0.033 |
| 278 |
246380_at |
AT1G57750
|
CYP96A15, cytochrome P450, family 96, subfamily A, polypeptide 15, MAH1, MID-CHAIN ALKANE HYDROXYLASE 1 |
0.163 |
0.052 |
| 279 |
260028_at |
AT1G29980
|
unknown |
0.163 |
0.007 |
| 280 |
266848_at |
AT2G25950
|
unknown |
0.163 |
0.056 |
| 281 |
251037_at |
AT5G02100
|
ORP3A, OSBP(OXYSTEROL BINDING PROTEIN)-RELATED PROTEIN 3A, UNE18, UNFERTILIZED EMBRYO SAC 18 |
0.162 |
-0.000 |
| 282 |
255433_at |
AT4G03210
|
XTH9, xyloglucan endotransglucosylase/hydrolase 9 |
0.161 |
-0.152 |
| 283 |
251927_at |
AT3G53990
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.160 |
0.009 |
| 284 |
259061_at |
AT3G07410
|
AtRABA5b, RAB GTPase homolog A5B, RABA5b, RAB GTPase homolog A5B |
0.159 |
-0.114 |
| 285 |
247509_at |
AT5G62020
|
AT-HSFB2A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR B2A, HSFB2A, heat shock transcription factor B2A |
0.159 |
0.027 |
| 286 |
246906_at |
AT5G25475
|
[AP2/B3-like transcriptional factor family protein] |
0.159 |
-0.068 |
| 287 |
260401_at |
AT1G69840
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
0.158 |
-0.060 |
| 288 |
257216_at |
AT3G14990
|
AtDJ1A, DJ-1 homolog A, DJ-1a, DJ1A, DJ-1 homolog A |
0.156 |
0.048 |
| 289 |
255625_at |
AT4G01120
|
ATBZIP54, BASIC REGION/LEUCINE ZIPPER MOTIF 5, GBF2, G-box binding factor 2 |
0.156 |
0.022 |
| 290 |
260910_at |
AT1G02690
|
IMPA-6, importin alpha isoform 6 |
0.156 |
-0.078 |
| 291 |
245557_at |
AT4G15410
|
PUX5, serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B prime gamma |
0.156 |
0.039 |
| 292 |
264338_at |
AT1G70300
|
KUP6, K+ uptake permease 6 |
0.154 |
0.170 |
| 293 |
254580_at |
AT4G19390
|
[Uncharacterised protein family (UPF0114)] |
0.154 |
0.166 |
| 294 |
265383_at |
AT2G16780
|
MSI02, MSI2, MULTICOPY SUPPRESSOR OF IRA1 2, NFC02, NFC2, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP C 2 |
0.154 |
-0.046 |
| 295 |
252427_at |
AT3G47640
|
PYE, POPEYE |
0.153 |
0.224 |
| 296 |
251846_at |
AT3G54560
|
HTA11, histone H2A 11 |
0.152 |
-0.054 |
| 297 |
266237_at |
AT2G29540
|
ATRPAC14, ATRPC14, RNApolymerase 14 kDa subunit, RPC14, RNApolymerase 14 kDa subunit |
0.151 |
-0.049 |
| 298 |
261859_at |
AT1G50490
|
UBC20, ubiquitin-conjugating enzyme 20 |
0.151 |
-0.125 |
| 299 |
258759_at |
AT3G10800
|
BZIP28 |
0.150 |
0.060 |
| 300 |
252322_at |
AT3G48550
|
unknown |
0.150 |
-0.045 |
| 301 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-0.528 |
-0.020 |
| 302 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
-0.460 |
0.108 |
| 303 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
-0.437 |
0.142 |
| 304 |
246597_at |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
-0.399 |
0.269 |
| 305 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
-0.395 |
-0.093 |
| 306 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
-0.365 |
-0.016 |
| 307 |
256619_at |
AT3G24460
|
[Serinc-domain containing serine and sphingolipid biosynthesis protein] |
-0.365 |
0.071 |
| 308 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.354 |
-0.155 |
| 309 |
247055_at |
AT5G66740
|
unknown |
-0.351 |
0.036 |
| 310 |
266078_at |
AT2G40670
|
ARR16, response regulator 16, RR16, response regulator 16 |
-0.347 |
0.033 |
| 311 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
-0.339 |
0.007 |
| 312 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.330 |
0.061 |
| 313 |
249614_at |
AT5G37300
|
WSD1 |
-0.329 |
0.054 |
| 314 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
-0.314 |
0.270 |
| 315 |
262212_at |
AT1G74890
|
ARR15, response regulator 15 |
-0.297 |
0.068 |
| 316 |
261046_at |
AT1G01390
|
[UDP-Glycosyltransferase superfamily protein] |
-0.285 |
0.076 |
| 317 |
252859_at |
AT4G39780
|
[Integrase-type DNA-binding superfamily protein] |
-0.281 |
0.181 |
| 318 |
256926_at |
AT3G22540
|
unknown |
-0.268 |
-0.027 |
| 319 |
246700_at |
AT5G28030
|
DES1, L-cysteine desulfhydrase 1 |
-0.264 |
0.019 |
| 320 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
-0.263 |
-0.030 |
| 321 |
262883_at |
AT1G64780
|
AMT1;2, ammonium transporter 1;2, ATAMT1;2, ammonium transporter 1;2 |
-0.257 |
-0.060 |
| 322 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
-0.256 |
0.109 |
| 323 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.253 |
-0.089 |
| 324 |
247684_at |
AT5G59670
|
[Leucine-rich repeat protein kinase family protein] |
-0.251 |
-0.022 |
| 325 |
247878_at |
AT5G57760
|
unknown |
-0.249 |
0.022 |
| 326 |
252300_at |
AT3G49160
|
[pyruvate kinase family protein] |
-0.244 |
0.288 |
| 327 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
-0.241 |
0.224 |
| 328 |
246069_at |
AT5G20220
|
[zinc knuckle (CCHC-type) family protein] |
-0.240 |
0.037 |
| 329 |
255264_at |
AT4G05170
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.238 |
-0.002 |
| 330 |
261350_at |
AT1G79770
|
unknown |
-0.238 |
0.033 |
| 331 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
-0.238 |
-0.071 |
| 332 |
250696_at |
AT5G06790
|
unknown |
-0.236 |
-0.069 |
| 333 |
253835_at |
AT4G27820
|
BGLU9, beta glucosidase 9 |
-0.235 |
0.144 |
| 334 |
263918_at |
AT2G36590
|
ATPROT3, PROLINE TRANSPORTER 3, ProT3, proline transporter 3 |
-0.234 |
0.068 |
| 335 |
257451_at |
AT1G05690
|
BT3, BTB and TAZ domain protein 3 |
-0.233 |
0.002 |
| 336 |
251066_at |
AT5G01880
|
DAFL2, DAF-Like gene 2 |
-0.232 |
0.235 |
| 337 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
-0.229 |
0.176 |
| 338 |
263486_at |
AT2G22200
|
[Integrase-type DNA-binding superfamily protein] |
-0.229 |
0.016 |
| 339 |
254683_at |
AT4G13800
|
unknown |
-0.226 |
-0.004 |
| 340 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
-0.225 |
-0.103 |
| 341 |
245616_at |
AT4G14480
|
[Protein kinase superfamily protein] |
-0.222 |
0.021 |
| 342 |
262737_at |
AT1G28560
|
SRD2, SHOOT REDIFFERENTIATION DEFECTIVE 2 |
-0.219 |
0.013 |
| 343 |
248246_at |
AT5G53200
|
TRY, TRIPTYCHON |
-0.218 |
0.009 |
| 344 |
245676_at |
AT1G56670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.216 |
-0.061 |
| 345 |
252917_at |
AT4G38960
|
BBX19, B-box domain protein 19 |
-0.215 |
0.027 |
| 346 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
-0.214 |
0.011 |
| 347 |
259871_at |
AT1G76800
|
[Vacuolar iron transporter (VIT) family protein] |
-0.214 |
0.022 |
| 348 |
265962_at |
AT2G37460
|
UMAMIT12, Usually multiple acids move in and out Transporters 12 |
-0.210 |
-0.308 |
| 349 |
245349_at |
AT4G16690
|
ATMES16, ARABIDOPSIS THALIANA METHYL ESTERASE 16, MES16, methyl esterase 16 |
-0.210 |
0.010 |
| 350 |
256379_at |
AT1G66840
|
PMI2, plastid movement impaired 2, WEB2, WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 2 |
-0.209 |
0.005 |
| 351 |
252010_at |
AT3G52740
|
unknown |
-0.208 |
0.145 |
| 352 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
-0.207 |
0.017 |
| 353 |
260603_at |
AT1G55960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.207 |
0.054 |
| 354 |
266464_at |
AT2G47800
|
ABCC4, ATP-binding cassette C4, ATMRP4, multidrug resistance-associated protein 4, EST3, MRP4, multidrug resistance-associated protein 4 |
-0.205 |
0.246 |
| 355 |
263115_at |
AT1G03055
|
AtD27, A. thaliana homolog of rice D27, D27, DWARF27 |
-0.204 |
0.045 |
| 356 |
250381_at |
AT5G11610
|
[Exostosin family protein] |
-0.204 |
0.097 |
| 357 |
267066_at |
AT2G41040
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.204 |
0.002 |
| 358 |
258315_at |
AT3G16175
|
[Thioesterase superfamily protein] |
-0.201 |
-0.001 |
| 359 |
267592_at |
AT2G39710
|
[Eukaryotic aspartyl protease family protein] |
-0.200 |
0.064 |
| 360 |
256386_at |
AT1G66540
|
[Cytochrome P450 superfamily protein] |
-0.200 |
0.083 |
| 361 |
254125_at |
AT4G24670
|
TAR2, tryptophan aminotransferase related 2 |
-0.198 |
-0.107 |
| 362 |
255016_at |
AT4G10120
|
ATSPS4F |
-0.198 |
0.030 |
| 363 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
-0.198 |
0.096 |
| 364 |
248889_at |
AT5G46230
|
unknown |
-0.197 |
-0.273 |
| 365 |
262811_at |
AT1G11700
|
unknown |
-0.197 |
0.027 |
| 366 |
258677_at |
AT3G08730
|
ATPK1, ARABIDOPSIS THALIANA PROTEIN-SERINE KINASE 1, ATPK6, ARABIDOPSIS THALIANA PROTEIN-SERINE KINASE 6, ATS6K1, PK1, protein-serine kinase 1, PK6, ROTEIN-SERINE KINASE 6, S6K1, P70 RIBOSOMAL S6 KINASE |
-0.196 |
0.081 |
| 367 |
258932_at |
AT3G10150
|
ATPAP16, PAP16, purple acid phosphatase 16 |
-0.195 |
0.077 |
| 368 |
263847_at |
AT2G36970
|
[UDP-Glycosyltransferase superfamily protein] |
-0.195 |
-0.013 |
| 369 |
266131_at |
AT2G45160
|
ATHAM1, ARABIDOPSIS THALIANA HAIRY MERISTEM 1, HAM1, HAIRY MERISTEM 1, LOM1, LOST MERISTEMS 1 |
-0.195 |
-0.013 |
| 370 |
264893_at |
AT1G23140
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.194 |
-0.015 |
| 371 |
249278_at |
AT5G41900
|
[alpha/beta-Hydrolases superfamily protein] |
-0.194 |
-0.028 |
| 372 |
261059_at |
AT1G01250
|
[Integrase-type DNA-binding superfamily protein] |
-0.194 |
0.045 |
| 373 |
259822_at |
AT1G66230
|
AtMYB20, myb domain protein 20, MYB20, myb domain protein 20 |
-0.192 |
0.055 |
| 374 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
-0.191 |
0.262 |
| 375 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.191 |
0.026 |
| 376 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
-0.190 |
0.183 |
| 377 |
250566_at |
AT5G08070
|
TCP17, TCP domain protein 17 |
-0.189 |
0.044 |
| 378 |
265573_at |
AT2G28200
|
[C2H2-type zinc finger family protein] |
-0.189 |
-0.254 |
| 379 |
245748_at |
AT1G51140
|
AKS1, ABA-responsive kinase substrate 1, FBH3, FLOWERING BHLH 3 |
-0.189 |
0.290 |
| 380 |
261618_at |
AT1G33110
|
[MATE efflux family protein] |
-0.188 |
0.240 |
| 381 |
250408_at |
AT5G10930
|
CIPK5, CBL-interacting protein kinase 5, SnRK3.24, SNF1-RELATED PROTEIN KINASE 3.24 |
-0.188 |
0.007 |
| 382 |
260367_at |
AT1G69760
|
unknown |
-0.187 |
0.141 |
| 383 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
-0.187 |
0.102 |
| 384 |
261836_at |
AT1G16090
|
WAKL7, wall associated kinase-like 7 |
-0.187 |
-0.006 |
| 385 |
266946_at |
AT2G18890
|
[Protein kinase superfamily protein] |
-0.186 |
0.148 |
| 386 |
249727_at |
AT5G35490
|
ATMRU1, ARABIDOPSIS MTO 1 RESPONDING UP 1, MRU1, MTO 1 RESPONDING UP 1 |
-0.185 |
0.012 |
| 387 |
254235_at |
AT4G23750
|
CRF2, cytokinin response factor 2, TMO3, TARGET OF MONOPTEROS 3 |
-0.185 |
0.087 |
| 388 |
257314_at |
AT3G26590
|
[MATE efflux family protein] |
-0.185 |
0.052 |
| 389 |
248890_at |
AT5G46270
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.184 |
0.003 |
| 390 |
267010_at |
AT2G39250
|
SNZ, SCHNARCHZAPFEN |
-0.184 |
0.068 |
| 391 |
256874_at |
AT3G26320
|
CYP71B36, cytochrome P450, family 71, subfamily B, polypeptide 36 |
-0.181 |
0.076 |
| 392 |
267637_at |
AT2G42190
|
unknown |
-0.181 |
-0.298 |
| 393 |
257710_at |
AT3G27350
|
unknown |
-0.180 |
0.042 |
| 394 |
246142_at |
AT5G19970
|
unknown |
-0.179 |
-0.234 |
| 395 |
264444_at |
AT1G27360
|
SPL11, squamosa promoter-like 11 |
-0.179 |
0.010 |
| 396 |
260170_at |
AT1G71890
|
ATSUC5, SUCROSE-PROTON SYMPORTER 5, SUC5 |
-0.179 |
0.034 |
| 397 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-0.178 |
0.202 |
| 398 |
248819_at |
AT5G47050
|
[SBP (S-ribonuclease binding protein) family protein] |
-0.178 |
-0.029 |
| 399 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
-0.178 |
-0.053 |
| 400 |
261068_at |
AT1G07450
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.178 |
-0.021 |
| 401 |
252414_at |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
-0.177 |
0.116 |
| 402 |
258409_at |
AT3G17640
|
[Leucine-rich repeat (LRR) family protein] |
-0.176 |
0.024 |
| 403 |
262281_at |
AT1G68570
|
AtNPF3.1, NPF3.1, NRT1/ PTR family 3.1 |
-0.175 |
0.332 |
| 404 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
-0.172 |
0.055 |
| 405 |
264200_at |
AT1G22650
|
A/N-InvD, alkaline/neutral invertase D |
-0.172 |
-0.079 |
| 406 |
253496_at |
AT4G31870
|
ATGPX7, GLUTATHIONE PEROXIDASE 7, GPX7, glutathione peroxidase 7 |
-0.172 |
0.049 |
| 407 |
250049_at |
AT5G17780
|
[alpha/beta-Hydrolases superfamily protein] |
-0.171 |
-0.019 |
| 408 |
256873_at |
AT3G26310
|
CYP71B35, cytochrome P450, family 71, subfamily B, polypeptide 35 |
-0.171 |
0.011 |
| 409 |
248306_at |
AT5G52830
|
ATWRKY27, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 27, WRKY27, WRKY DNA-binding protein 27 |
-0.170 |
0.084 |
| 410 |
250744_at |
AT5G05840
|
unknown |
-0.170 |
0.033 |
| 411 |
257959_at |
AT3G25560
|
AtNIK2, NIK2, NSP-interacting kinase 2 |
-0.170 |
-0.088 |
| 412 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
-0.170 |
0.072 |
| 413 |
249336_at |
AT5G41070
|
DRB5, dsRNA-binding protein 5 |
-0.170 |
0.036 |
| 414 |
262526_at |
AT1G17050
|
AtSPS2, SPS2, solanesyl diphosphate synthase 2 |
-0.170 |
0.208 |
| 415 |
266391_at |
AT2G41290
|
SSL2, strictosidine synthase-like 2 |
-0.170 |
-0.076 |
| 416 |
261651_at |
AT1G27760
|
ATSAT32, SALT-TOLERANCE 32, SAT32, SALT-TOLERANCE 32 |
-0.170 |
0.125 |
| 417 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
-0.169 |
-0.068 |
| 418 |
259348_at |
AT3G03770
|
[Leucine-rich repeat protein kinase family protein] |
-0.168 |
0.141 |
| 419 |
251301_at |
AT3G61880
|
CYP78A9, cytochrome p450 78a9 |
-0.168 |
0.133 |
| 420 |
262464_at |
AT1G50280
|
[Phototropic-responsive NPH3 family protein] |
-0.168 |
0.021 |
| 421 |
256681_at |
AT3G52340
|
ATSPP2, SUCROSE-PHOSPHATASE 2, SPP2, sucrose-6F-phosphate phosphohydrolase 2 |
-0.168 |
0.295 |
| 422 |
258807_at |
AT3G04030
|
MYR2 |
-0.167 |
0.016 |
| 423 |
265842_at |
AT2G35700
|
ATERF38, ERF FAMILY PROTEIN 38, ERF38, ERF family protein 38 |
-0.167 |
0.011 |
| 424 |
250161_at |
AT5G15240
|
[Transmembrane amino acid transporter family protein] |
-0.166 |
0.077 |
| 425 |
265272_at |
AT2G28350
|
ARF10, auxin response factor 10 |
-0.166 |
-0.023 |
| 426 |
249798_at |
AT5G23730
|
EFO2, EARLY FLOWERING BY OVEREXPRESSION 2, RUP2, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 2 |
-0.166 |
0.017 |
| 427 |
251020_at |
AT5G02270
|
ABCI20, ATP-binding cassette I20, ATNAP9, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN 9, NAP9, non-intrinsic ABC protein 9 |
-0.164 |
-0.048 |
| 428 |
264083_at |
AT2G31230
|
ATERF15, ethylene-responsive element binding factor 15, ERF15, ethylene-responsive element binding factor 15 |
-0.164 |
0.037 |
| 429 |
260491_at |
AT1G51440
|
DALL2, DAD1-Like Lipase 2 |
-0.164 |
0.077 |
| 430 |
265203_at |
AT2G36630
|
[Sulfite exporter TauE/SafE family protein] |
-0.164 |
0.067 |
| 431 |
257999_at |
AT3G27540
|
[beta-1,4-N-acetylglucosaminyltransferase family protein] |
-0.164 |
0.050 |
| 432 |
245593_at |
AT4G14550
|
IAA14, indole-3-acetic acid inducible 14, SLR, SOLITARY ROOT |
-0.163 |
-0.062 |
| 433 |
249011_at |
AT5G44670
|
GALS2, galactan synthase 2 |
-0.163 |
-0.068 |
| 434 |
245723_at |
AT1G73400
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.162 |
0.003 |
| 435 |
258982_at |
AT3G08870
|
LecRK-VI.1, L-type lectin receptor kinase VI.1 |
-0.162 |
0.031 |
| 436 |
252374_at |
AT3G48100
|
ARR5, response regulator 5, ATRR2, ARABIDOPSIS THALIANA RESPONSE REGULATOR 2, IBC6, INDUCED BY CYTOKININ 6, RR5, response regulator 5 |
-0.161 |
-0.445 |
| 437 |
250499_at |
AT5G09730
|
ATBX3, ATBXL3, BETA-XYLOSIDASE 3, BX3, BXL3, beta-xylosidase 3, XYL3 |
-0.161 |
-0.027 |
| 438 |
256674_at |
AT3G52360
|
unknown |
-0.161 |
0.028 |
| 439 |
249059_at |
AT5G44530
|
[Subtilase family protein] |
-0.161 |
-0.009 |
| 440 |
266778_at |
AT2G29090
|
CYP707A2, cytochrome P450, family 707, subfamily A, polypeptide 2 |
-0.161 |
0.059 |
| 441 |
256819_at |
AT3G21390
|
[Mitochondrial substrate carrier family protein] |
-0.161 |
-0.053 |
| 442 |
262935_at |
AT1G79410
|
AtOCT5, organic cation/carnitine transporter5, OCT5, organic cation/carnitine transporter5 |
-0.160 |
-0.057 |
| 443 |
260059_at |
AT1G78090
|
ATTPPB, Arabidopsis thaliana trehalose-6-phosphate phosphatase B, TPPB, trehalose-6-phosphate phosphatase B |
-0.160 |
-0.143 |
| 444 |
254145_at |
AT4G24700
|
unknown |
-0.160 |
-0.038 |
| 445 |
245152_at |
AT2G47490
|
ATNDT1, ARABIDOPSIS THALIANA NAD+ TRANSPORTER 1, NDT1, NAD+ transporter 1 |
-0.160 |
-0.053 |
| 446 |
256528_at |
AT1G66140
|
ZFP4, zinc finger protein 4 |
-0.160 |
-0.040 |
| 447 |
254133_at |
AT4G24810
|
[Protein kinase superfamily protein] |
-0.160 |
0.058 |
| 448 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.159 |
0.089 |
| 449 |
257545_at |
AT3G23200
|
[Uncharacterised protein family (UPF0497)] |
-0.159 |
0.088 |
| 450 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
-0.159 |
0.056 |
| 451 |
263252_at |
AT2G31380
|
BBX25, B-box domain protein 25, STH, salt tolerance homologue |
-0.157 |
-0.003 |
| 452 |
264318_at |
AT1G04220
|
KCS2, 3-ketoacyl-CoA synthase 2 |
-0.157 |
0.097 |
| 453 |
248607_at |
AT5G49480
|
ATCP1, Ca2+-binding protein 1, CP1, Ca2+-binding protein 1 |
-0.157 |
0.104 |
| 454 |
253579_at |
AT4G30610
|
BRS1, BRI1 SUPPRESSOR 1, SCPL24, SERINE CARBOXYPEPTIDASE 24 PRECURSOR |
-0.157 |
-0.031 |
| 455 |
264909_at |
AT2G17300
|
unknown |
-0.157 |
-0.202 |
| 456 |
246566_at |
AT5G14940
|
[Major facilitator superfamily protein] |
-0.156 |
0.106 |
| 457 |
266007_at |
AT2G37380
|
MAKR3, MEMBRANE-ASSOCIATED KINASE REGULATOR 3 |
-0.156 |
0.031 |
| 458 |
263122_at |
AT1G78510
|
AtSPS1, SPS1, solanesyl diphosphate synthase 1 |
-0.156 |
0.111 |
| 459 |
253100_at |
AT4G37400
|
CYP81F3, cytochrome P450, family 81, subfamily F, polypeptide 3 |
-0.156 |
-0.063 |
| 460 |
251110_at |
AT5G01260
|
[Carbohydrate-binding-like fold] |
-0.156 |
0.074 |
| 461 |
265892_at |
AT2G15020
|
unknown |
-0.156 |
0.007 |
| 462 |
253048_at |
AT4G37560
|
[Acetamidase/Formamidase family protein] |
-0.155 |
-0.014 |
| 463 |
252073_at |
AT3G51750
|
unknown |
-0.155 |
0.920 |
| 464 |
245936_at |
AT5G19850
|
[alpha/beta-Hydrolases superfamily protein] |
-0.155 |
0.126 |
| 465 |
247726_at |
AT5G59430
|
ATTRP1, TELOMERE REPEAT BINDING PROTEIN 1, TRP1, telomeric repeat binding protein 1 |
-0.155 |
0.165 |
| 466 |
260774_at |
AT1G78290
|
SNRK2-8, SNF1-RELATED PROTEIN KINASE 2-8, SNRK2.8, SNF1-RELATED PROTEIN KINASE 2.8, SRK2C, SNF1-RELATED PROTEIN KINASE 2C |
-0.155 |
-0.125 |
| 467 |
257543_at |
AT3G28960
|
[Transmembrane amino acid transporter family protein] |
-0.154 |
-0.020 |
| 468 |
246001_at |
AT5G20790
|
unknown |
-0.154 |
0.061 |
| 469 |
249309_at |
AT5G41410
|
BEL1, BELL 1 |
-0.154 |
0.024 |
| 470 |
266362_at |
AT2G32430
|
[Galactosyltransferase family protein] |
-0.154 |
-0.008 |
| 471 |
260156_at |
AT1G52880
|
ANAC018, Arabidopsis NAC domain containing protein 18, ATNAM, NAM, NO APICAL MERISTEM, NARS2, NAC-REGULATED SEED MORPHOLOGY 2 |
-0.153 |
0.347 |
| 472 |
251024_at |
AT5G02180
|
[Transmembrane amino acid transporter family protein] |
-0.153 |
-0.119 |
| 473 |
251727_at |
AT3G56290
|
unknown |
-0.153 |
-0.133 |
| 474 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
-0.152 |
-0.144 |
| 475 |
245965_at |
AT5G19730
|
[Pectin lyase-like superfamily protein] |
-0.152 |
0.138 |
| 476 |
261958_at |
AT1G64500
|
[Glutaredoxin family protein] |
-0.152 |
0.068 |
| 477 |
247980_at |
AT5G56860
|
GATA21, GATA TRANSCRIPTION FACTOR 21, GNC, GATA, nitrate-inducible, carbon metabolism-involved |
-0.152 |
-0.014 |
| 478 |
262136_at |
AT1G77850
|
ARF17, auxin response factor 17 |
-0.152 |
0.013 |
| 479 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
-0.152 |
0.139 |
| 480 |
247522_at |
AT5G61340
|
unknown |
-0.152 |
-0.065 |
| 481 |
256096_at |
AT1G13650
|
unknown |
-0.151 |
0.072 |
| 482 |
249464_at |
AT5G39710
|
EMB2745, EMBRYO DEFECTIVE 2745 |
-0.151 |
0.027 |
| 483 |
253692_at |
AT4G29720
|
ATPAO5, polyamine oxidase 5, PAO5, polyamine oxidase 5 |
-0.151 |
-0.253 |
| 484 |
260773_at |
AT1G78440
|
ATGA2OX1, Arabidopsis thaliana gibberellin 2-oxidase 1, GA2OX1, gibberellin 2-oxidase 1 |
-0.151 |
0.055 |
| 485 |
266830_at |
AT2G22810
|
ACC4, 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE POLYPEPTIDE, ACS4, 1-aminocyclopropane-1-carboxylate synthase 4, ATACS4 |
-0.151 |
0.002 |
| 486 |
258905_at |
AT3G06390
|
[Uncharacterised protein family (UPF0497)] |
-0.150 |
-0.185 |
| 487 |
257801_at |
AT3G18750
|
ATWNK6, ARABIDOPSIS THALIANA WITH NO K 6, WNK6, with no lysine (K) kinase 6, ZIK5 |
-0.150 |
-0.133 |
| 488 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
-0.150 |
-0.029 |
| 489 |
253165_at |
AT4G35320
|
unknown |
-0.150 |
-0.184 |
| 490 |
253517_at |
AT4G31390
|
ABC1K1, ABC1-like kinase 1, ACDO1, ABC1-like kinase related to chlorophyll degradation and oxidative stress 1, AtACDO1, PGR6, PROTON GRADIENT REGULATION 6 |
-0.149 |
0.036 |
| 491 |
256302_at |
AT1G69526
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.149 |
-0.068 |
| 492 |
256894_at |
AT3G21870
|
CYCP2;1, cyclin p2;1 |
-0.148 |
-0.006 |
| 493 |
263168_at |
AT1G03020
|
[Thioredoxin superfamily protein] |
-0.148 |
0.024 |
| 494 |
252096_at |
AT3G51180
|
[Zinc finger C-x8-C-x5-C-x3-H type family protein] |
-0.148 |
-0.080 |
| 495 |
254024_at |
AT4G25780
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.147 |
-0.350 |
| 496 |
256735_at |
AT3G29400
|
ATEXO70E1, exocyst subunit exo70 family protein E1, EXO70E1, exocyst subunit exo70 family protein E1 |
-0.147 |
0.067 |
| 497 |
263919_at |
AT2G36470
|
unknown |
-0.147 |
0.165 |
| 498 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
-0.147 |
0.053 |
| 499 |
258419_at |
AT3G16670
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.147 |
0.080 |
| 500 |
252204_at |
AT3G50340
|
unknown |
-0.147 |
0.011 |
| 501 |
245724_at |
AT1G73390
|
[Endosomal targeting BRO1-like domain-containing protein] |
-0.147 |
0.279 |
| 502 |
248118_at |
AT5G55050
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.147 |
0.031 |
| 503 |
265734_at |
AT2G01260
|
unknown |
-0.147 |
-0.036 |
| 504 |
261776_at |
AT1G76190
|
SAUR56, SMALL AUXIN UPREGULATED RNA 56 |
-0.146 |
0.035 |
| 505 |
265726_at |
AT2G32010
|
CVL1, CVP2 like 1 |
-0.146 |
0.039 |
| 506 |
254159_at |
AT4G24240
|
ATWRKY7, WRKY7, WRKY DNA-binding protein 7 |
-0.146 |
0.039 |
| 507 |
261845_at |
AT1G15960
|
ATNRAMP6, NRAMP6, NRAMP metal ion transporter 6 |
-0.146 |
-0.031 |
| 508 |
263796_at |
AT2G24540
|
AFR, ATTENUATED FAR-RED RESPONSE |
-0.146 |
-0.039 |
| 509 |
248440_at |
AT5G51260
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.146 |
0.180 |
| 510 |
251154_at |
AT3G63110
|
ATIPT3, isopentenyltransferase 3, IPT3, isopentenyltransferase 3 |
-0.146 |
-0.011 |
| 511 |
261758_at |
AT1G08250
|
ADT6, arogenate dehydratase 6, AtADT6, Arabidopsis thaliana arogenate dehydratase 6 |
-0.145 |
0.029 |
| 512 |
264783_at |
AT1G08650
|
ATPPCK1, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 1, PPCK1, phosphoenolpyruvate carboxylase kinase 1 |
-0.145 |
0.112 |
| 513 |
259209_at |
AT3G09160
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.145 |
-0.008 |
| 514 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
-0.145 |
0.355 |
| 515 |
252911_at |
AT4G39510
|
CYP96A12, cytochrome P450, family 96, subfamily A, polypeptide 12 |
-0.145 |
0.033 |
| 516 |
257147_at |
AT3G27270
|
[TRAM, LAG1 and CLN8 (TLC) lipid-sensing domain containing protein] |
-0.144 |
-0.011 |
| 517 |
259359_at |
AT1G13460
|
[Protein phosphatase 2A regulatory B subunit family protein] |
-0.144 |
-0.088 |
| 518 |
250665_at |
AT5G06980
|
LNK4, night light-inducible and clock-regulated 4 |
-0.144 |
0.231 |
| 519 |
248080_at |
AT5G55380
|
[MBOAT (membrane bound O-acyl transferase) family protein] |
-0.144 |
-0.038 |
| 520 |
262552_at |
AT1G31350
|
KUF1, KAR-UP F-box 1 |
-0.143 |
0.089 |
| 521 |
263241_at |
AT2G16500
|
ADC1, arginine decarboxylase 1, ARGDC, ARGDC1, SPE1 |
-0.143 |
-0.098 |
| 522 |
264956_at |
AT1G76990
|
ACR3, ACT domain repeat 3 |
-0.143 |
0.033 |
| 523 |
263386_at |
AT2G40150
|
TBL28, TRICHOME BIREFRINGENCE-LIKE 28 |
-0.143 |
-0.294 |
| 524 |
248756_at |
AT5G47560
|
ATSDAT, ATTDT, TONOPLAST DICARBOXYLATE TRANSPORTER, TDT, tonoplast dicarboxylate transporter |
-0.143 |
0.212 |
| 525 |
264436_at |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
-0.143 |
0.432 |
| 526 |
258119_at |
AT3G14720
|
ATMPK19, ARABIDOPSIS THALIANA MAP KINASE 19, MPK19, MAP kinase 19 |
-0.142 |
0.079 |
| 527 |
260064_at |
AT1G73730
|
AtEIL3, ATSLIM, ARABIDOPSIS THALIANA SULFUR LIMITATION 1, EIL3, ETHYLENE-INSENSITIVE3-like 3, SLIM1, SULFUR LIMITATION 1 |
-0.142 |
0.116 |
| 528 |
258023_at |
AT3G19450
|
ATCAD4, CAD, CAD-C, CAD4, CINNAMYL ALCOHOL DEHYDROGENASE 4 |
-0.142 |
0.006 |
| 529 |
251890_at |
AT3G54220
|
SCR, SCARECROW, SGR1, SHOOT GRAVITROPISM 1 |
-0.141 |
-0.040 |
| 530 |
255877_at |
AT2G40460
|
[Major facilitator superfamily protein] |
-0.141 |
-0.020 |
| 531 |
264692_at |
AT1G70000
|
[myb-like transcription factor family protein] |
-0.141 |
0.013 |
| 532 |
259856_at |
AT1G68440
|
unknown |
-0.141 |
0.069 |
| 533 |
261319_at |
AT1G53090
|
SPA4, SPA1-related 4 |
-0.141 |
0.055 |
| 534 |
253814_at |
AT4G28290
|
unknown |
-0.141 |
0.115 |
| 535 |
266809_at |
AT2G29970
|
SMXL7, SMAX1-like 7 |
-0.140 |
0.102 |
| 536 |
246462_at |
AT5G16940
|
[carbon-sulfur lyases] |
-0.139 |
-0.096 |
| 537 |
265339_at |
AT2G18230
|
AtPPa2, pyrophosphorylase 2, PPa2, pyrophosphorylase 2 |
-0.139 |
0.081 |
| 538 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
-0.139 |
-0.022 |
| 539 |
254293_at |
AT4G23060
|
IQD22, IQ-domain 22 |
-0.139 |
0.120 |
| 540 |
252482_at |
AT3G46670
|
UGT76E11, UDP-glucosyl transferase 76E11 |
-0.138 |
0.182 |
| 541 |
257850_at |
AT3G13065
|
SRF4, STRUBBELIG-receptor family 4 |
-0.138 |
0.029 |
| 542 |
257300_at |
AT3G28080
|
UMAMIT47, Usually multiple acids move in and out Transporters 47 |
-0.138 |
0.016 |
| 543 |
245197_at |
AT1G67800
|
[Copine (Calcium-dependent phospholipid-binding protein) family] |
-0.138 |
0.098 |
| 544 |
245416_at |
AT4G17350
|
unknown |
-0.138 |
0.183 |
| 545 |
264102_at |
AT1G79270
|
ECT8, evolutionarily conserved C-terminal region 8 |
-0.137 |
0.410 |
| 546 |
255129_at |
AT4G08290
|
UMAMIT20, Usually multiple acids move in and out Transporters 20 |
-0.137 |
-0.025 |
| 547 |
254693_at |
AT4G17880
|
MYC4 |
-0.137 |
-0.128 |
| 548 |
263677_at |
AT1G04520
|
PDLP2, plasmodesmata-located protein 2 |
-0.137 |
-0.085 |
| 549 |
253264_at |
AT4G33950
|
ATOST1, OPEN STOMATA 1, OST1, OPEN STOMATA 1, P44, SNRK2-6, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-6, SNRK2.6, SNF1-RELATED PROTEIN KINASE 2.6, SRK2E |
-0.137 |
0.051 |
| 550 |
253040_at |
AT4G37800
|
XTH7, xyloglucan endotransglucosylase/hydrolase 7 |
-0.137 |
0.058 |
| 551 |
252661_at |
AT3G44450
|
unknown |
-0.137 |
0.004 |
| 552 |
253518_at |
AT4G31400
|
AtCTF7, CTF7, CHROMOSOME TRANSMISSION FIDELITY 7, ECO1 |
-0.136 |
-0.035 |
| 553 |
265939_at |
AT2G19650
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.136 |
-0.031 |
| 554 |
260979_at |
AT1G53510
|
ATMPK18, ARABIDOPSIS THALIANA MAP KINASE 18, MPK18, mitogen-activated protein kinase 18 |
-0.136 |
0.026 |
| 555 |
260309_at |
AT1G70580
|
AOAT2, alanine-2-oxoglutarate aminotransferase 2, GGT2, GLUTAMATE:GLYOXYLATE AMINOTRANSFERASE 2 |
-0.136 |
-0.095 |
| 556 |
246966_at |
AT5G24850
|
CRY3, cryptochrome 3 |
-0.136 |
0.081 |
| 557 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
-0.136 |
0.035 |
| 558 |
253411_at |
AT4G32980
|
ATH1, homeobox gene 1 |
-0.136 |
0.029 |
| 559 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
-0.136 |
0.031 |
| 560 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
-0.136 |
0.430 |
| 561 |
248039_at |
AT5G55950
|
[Nucleotide/sugar transporter family protein] |
-0.135 |
-0.007 |
| 562 |
253474_at |
AT4G32270
|
[Ubiquitin-like superfamily protein] |
-0.135 |
-0.086 |
| 563 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
-0.135 |
0.135 |
| 564 |
245277_at |
AT4G15550
|
IAGLU, indole-3-acetate beta-D-glucosyltransferase |
-0.135 |
0.093 |
| 565 |
264738_at |
AT1G62250
|
unknown |
-0.135 |
0.015 |
| 566 |
252363_at |
AT3G48460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.135 |
0.086 |
| 567 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
-0.135 |
0.055 |
| 568 |
264998_at |
AT1G67330
|
unknown |
-0.135 |
-0.169 |
| 569 |
254413_at |
AT4G21440
|
ATM4, A. THALIANA MYB 4, ATMYB102, MYB-like 102, MYB102, MYB102, MYB-like 102 |
-0.134 |
0.151 |
| 570 |
247533_at |
AT5G61570
|
[Protein kinase superfamily protein] |
-0.134 |
-0.115 |
| 571 |
266760_at |
AT2G46870
|
NGA1, NGATHA1 |
-0.134 |
0.060 |
| 572 |
250985_at |
AT5G02830
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.134 |
-0.017 |
| 573 |
260076_at |
AT1G73630
|
[EF hand calcium-binding protein family] |
-0.134 |
-0.049 |
| 574 |
250647_at |
AT5G06770
|
[KH domain-containing protein / zinc finger (CCCH type) family protein] |
-0.134 |
0.013 |
| 575 |
253052_at |
AT4G37310
|
CYP81H1, cytochrome P450, family 81, subfamily H, polypeptide 1 |
-0.134 |
0.026 |
| 576 |
247714_at |
AT5G59340
|
WOX2, WUSCHEL related homeobox 2 |
-0.133 |
0.071 |
| 577 |
259793_at |
AT1G64380
|
[Integrase-type DNA-binding superfamily protein] |
-0.133 |
0.479 |
| 578 |
246510_at |
AT5G15410
|
ATCNGC2, CYCLIC NUCLEOTIDE-GATED CHANNEL 2, CNGC2, CYCLIC NUCLEOTIDE GATED CHANNEL 2, DND1, DEFENSE NO DEATH 1 |
-0.133 |
0.048 |
| 579 |
260583_x_at |
AT2G47220
|
unknown |
-0.133 |
-0.055 |
| 580 |
246120_at |
AT5G20360
|
Phox3, Phox3 |
-0.133 |
0.056 |
| 581 |
245946_at |
AT5G19580
|
[glyoxal oxidase-related protein] |
-0.133 |
-0.111 |
| 582 |
263433_at |
AT2G22240
|
ATIPS2, INOSITOL 3-PHOSPHATE SYNTHASE 2, ATMIPS2, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 2, MIPS2, myo-inositol-1-phosphate synthase 2 |
-0.133 |
0.148 |
| 583 |
262290_at |
AT1G70985
|
[hydroxyproline-rich glycoprotein family protein] |
-0.133 |
0.074 |
| 584 |
255031_at |
AT4G09490
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
-0.133 |
0.019 |
| 585 |
263674_at |
AT2G04790
|
unknown |
-0.133 |
0.034 |
| 586 |
258520_at |
AT3G06710
|
unknown |
-0.133 |
-0.040 |
| 587 |
253055_at |
AT4G37630
|
CYCD5;1, cyclin d5;1 |
-0.132 |
-0.041 |
| 588 |
245528_at |
AT4G15530
|
PPDK, pyruvate orthophosphate dikinase |
-0.132 |
0.053 |
| 589 |
250859_at |
AT5G04660
|
CYP77A4, cytochrome P450, family 77, subfamily A, polypeptide 4 |
-0.132 |
0.237 |
| 590 |
248261_at |
AT5G53280
|
PDV1, PLASTID DIVISION1 |
-0.132 |
-0.034 |
| 591 |
251869_at |
AT3G54500
|
LNK2, night light-inducible and clock-regulated 2 |
-0.132 |
0.051 |
| 592 |
257253_at |
AT3G24190
|
[Protein kinase superfamily protein] |
-0.132 |
0.088 |
| 593 |
254689_at |
AT4G13710
|
[Pectin lyase-like superfamily protein] |
-0.132 |
0.080 |
| 594 |
258184_at |
AT3G21510
|
AHP1, histidine-containing phosphotransmitter 1 |
-0.132 |
0.069 |
| 595 |
245601_at |
AT4G14240
|
unknown |
-0.132 |
-0.049 |
| 596 |
263777_at |
AT2G46450
|
ATCNGC12, cyclic nucleotide-gated channel 12, CNGC12, cyclic nucleotide-gated channel 12 |
-0.131 |
0.025 |
| 597 |
256713_at |
AT2G34060
|
[Peroxidase superfamily protein] |
-0.131 |
0.015 |
| 598 |
263887_at |
AT2G36980
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.131 |
0.022 |
| 599 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
-0.131 |
0.118 |
| 600 |
267070_at |
AT2G41000
|
[Chaperone DnaJ-domain superfamily protein] |
-0.131 |
0.016 |