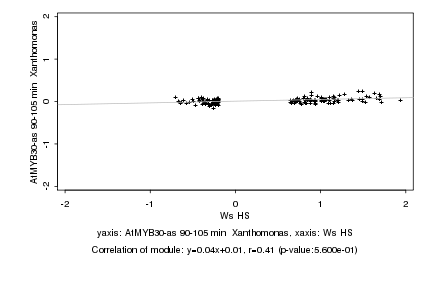
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Ws HS |
Log2 signal ratio
AtMYB30-as 90-105 min Xanthomonas |
| 1 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
1.933 |
0.046 |
| 2 |
262307_at |
AT1G71000
|
[Chaperone DnaJ-domain superfamily protein] |
1.711 |
-0.008 |
| 3 |
260978_at |
AT1G53540
|
[HSP20-like chaperones superfamily protein] |
1.692 |
0.130 |
| 4 |
253884_at |
AT4G27670
|
HSP21, heat shock protein 21 |
1.691 |
0.188 |
| 5 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
1.688 |
0.056 |
| 6 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
1.652 |
0.090 |
| 7 |
258695_at |
AT3G09640
|
APX1B, ASCORBATE PEROXIDASE 1B, APX2, ascorbate peroxidase 2 |
1.622 |
0.211 |
| 8 |
254059_at |
AT4G25200
|
ATHSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6, HSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6 |
1.572 |
0.108 |
| 9 |
249575_at |
AT5G37670
|
[HSP20-like chaperones superfamily protein] |
1.527 |
0.132 |
| 10 |
251166_at |
AT3G63350
|
AT-HSFA7B, HSFA7B, HEAT SHOCK TRANSCRIPTION FACTOR A7B |
1.525 |
-0.017 |
| 11 |
255811_at |
AT4G10250
|
ATHSP22.0 |
1.489 |
0.256 |
| 12 |
250624_at |
AT5G07330
|
unknown |
1.486 |
0.053 |
| 13 |
266841_at |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
1.485 |
0.018 |
| 14 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
1.447 |
0.053 |
| 15 |
247691_at |
AT5G59720
|
HSP18.2, heat shock protein 18.2 |
1.440 |
0.242 |
| 16 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
1.373 |
0.047 |
| 17 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
1.352 |
0.048 |
| 18 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
1.325 |
0.026 |
| 19 |
259913_at |
AT1G72660
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
1.277 |
0.168 |
| 20 |
257323_at |
ATMG01200
|
ORF294 |
1.217 |
0.149 |
| 21 |
261838_at |
AT1G16030
|
Hsp70b, heat shock protein 70B |
1.205 |
0.041 |
| 22 |
248215_at |
AT5G53680
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
1.202 |
-0.010 |
| 23 |
258827_at |
AT3G07150
|
unknown |
1.194 |
0.031 |
| 24 |
245243_at |
AT1G44414
|
unknown |
1.159 |
0.112 |
| 25 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
1.155 |
0.059 |
| 26 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
1.153 |
0.012 |
| 27 |
244902_at |
ATMG00650
|
NAD4L, NADH dehydrogenase subunit 4L |
1.148 |
0.126 |
| 28 |
256518_at |
AT1G66080
|
unknown |
1.143 |
-0.026 |
| 29 |
267190_at |
AT2G44170
|
ATNMT2, ARABIDOPSIS N-MYRISTOYLTRANSFERASE 2, NMT2, N-myristoyltransferase 2 |
1.143 |
0.084 |
| 30 |
244931_at |
ATMG00630
|
ORF110B |
1.108 |
-0.031 |
| 31 |
253859_at |
AT4G27657
|
unknown |
1.104 |
0.105 |
| 32 |
265675_at |
AT2G32120
|
HSP70T-2, heat-shock protein 70T-2 |
1.096 |
0.044 |
| 33 |
265242_at |
AT2G07705
|
unknown |
1.090 |
-0.029 |
| 34 |
250826_at |
AT5G05220
|
unknown |
1.058 |
0.068 |
| 35 |
254062_at |
AT4G25380
|
AtSAP10, Arabidopsis thaliana stress-associated protein 10, SAP10, stress-associated protein 10 |
1.050 |
0.042 |
| 36 |
255651_at |
AT4G00940
|
AtDOF4.1, DOF4.1, DNA binding with one finger 4.1, ITD1, INTERCELLULAR TRAFFICKING DOF 1 |
1.046 |
0.063 |
| 37 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
1.040 |
0.004 |
| 38 |
267391_at |
AT2G44480
|
BGLU17, beta glucosidase 17 |
1.040 |
0.011 |
| 39 |
263164_at |
AT1G03070
|
AtLFG4, LFG4, LIFEGUARD 4 |
1.039 |
0.061 |
| 40 |
267026_at |
AT2G38340
|
DREB19, dehydration response element-binding protein 19 |
1.031 |
0.001 |
| 41 |
251724_at |
AT3G56250
|
unknown |
1.024 |
0.061 |
| 42 |
244943_at |
ATMG00070
|
NAD9, NADH dehydrogenase subunit 9 |
1.010 |
0.097 |
| 43 |
267263_at |
AT2G23110
|
[Late embryogenesis abundant protein, group 6] |
1.009 |
0.022 |
| 44 |
254263_at |
AT4G23493
|
unknown |
1.006 |
0.092 |
| 45 |
246944_at |
AT5G25450
|
[Cytochrome bd ubiquinol oxidase, 14kDa subunit] |
1.003 |
0.028 |
| 46 |
244901_at |
ATMG00640
|
ORF25 |
0.953 |
0.129 |
| 47 |
264402_at |
AT2G25140
|
CLPB-M, CASEIN LYTIC PROTEINASE B-M, CLPB4, casein lytic proteinase B4, HSP98.7, HEAT SHOCK PROTEIN 98.7 |
0.938 |
0.035 |
| 48 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
0.934 |
-0.059 |
| 49 |
256285_at |
AT3G12510
|
[MADS-box family protein] |
0.933 |
-0.027 |
| 50 |
256363_at |
AT1G66510
|
[AAR2 protein family] |
0.930 |
0.003 |
| 51 |
244903_at |
ATMG00660
|
ORF149 |
0.925 |
0.030 |
| 52 |
260917_at |
AT1G02700
|
unknown |
0.923 |
0.007 |
| 53 |
244950_at |
ATMG00160
|
COX2, cytochrome oxidase 2 |
0.891 |
0.159 |
| 54 |
250162_at |
AT5G15250
|
ATFTSH6, FTSH6, FTSH protease 6 |
0.888 |
0.044 |
| 55 |
265660_at |
AT2G25470
|
AtRLP21, receptor like protein 21, RLP21, receptor like protein 21 |
0.884 |
0.219 |
| 56 |
245272_at |
AT4G17250
|
unknown |
0.879 |
0.011 |
| 57 |
263320_at |
AT2G47180
|
AtGolS1, galactinol synthase 1, GolS1, galactinol synthase 1 |
0.878 |
0.052 |
| 58 |
253293_at |
AT4G33905
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
0.873 |
-0.030 |
| 59 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
0.861 |
0.034 |
| 60 |
258406_at |
AT3G17611
|
ATRBL10, RHOMBOID-like protein 10, ATRBL14, RHOMBOID-like protein 14, RBL10, RHOMBOID-like protein 10, RBL14, RHOMBOID-like protein 14 |
0.845 |
0.037 |
| 61 |
245392_at |
AT4G15680
|
[Thioredoxin superfamily protein] |
0.842 |
0.081 |
| 62 |
244906_at |
ATMG00690
|
ORF240A |
0.839 |
0.057 |
| 63 |
261636_at |
AT1G50110
|
[D-aminoacid aminotransferase-like PLP-dependent enzymes superfamily protein] |
0.830 |
-0.034 |
| 64 |
247206_at |
AT5G64950
|
[Mitochondrial transcription termination factor family protein] |
0.821 |
0.029 |
| 65 |
253839_at |
AT4G27890
|
[HSP20-like chaperones superfamily protein] |
0.821 |
-0.025 |
| 66 |
259268_at |
AT3G01070
|
AtENODL16, ENODL16, early nodulin-like protein 16 |
0.809 |
0.138 |
| 67 |
258336_at |
AT3G16050
|
A37, ATPDX1.2, ARABIDOPSIS THALIANA PYRIDOXINE BIOSYNTHESIS 1.2, PDX1.2, pyridoxine biosynthesis 1.2 |
0.801 |
-0.013 |
| 68 |
253689_at |
AT4G29770
|
unknown |
0.790 |
0.092 |
| 69 |
261776_at |
AT1G76190
|
SAUR56, SMALL AUXIN UPREGULATED RNA 56 |
0.773 |
-0.047 |
| 70 |
257874_at |
AT3G17110
|
[pseudogene] |
0.756 |
-0.030 |
| 71 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
0.751 |
0.023 |
| 72 |
257336_at |
ATMG01410
|
ORF204, open reading frame 204 |
0.743 |
0.071 |
| 73 |
258794_at |
AT3G04710
|
TPR10, tetratricopeptide repeat 10 |
0.731 |
0.030 |
| 74 |
258603_at |
AT3G02990
|
ATHSFA1E, heat shock transcription factor A1E, HSFA1E, heat shock transcription factor A1E |
0.723 |
0.002 |
| 75 |
245979_at |
AT5G13150
|
ATEXO70C1, exocyst subunit exo70 family protein C1, EXO70C1, exocyst subunit exo70 family protein C1 |
0.722 |
0.078 |
| 76 |
253949_at |
AT4G26780
|
AR192, MGE2, mitochondrial GrpE 2 |
0.714 |
0.057 |
| 77 |
267462_at |
AT2G33735
|
[Chaperone DnaJ-domain superfamily protein] |
0.705 |
-0.017 |
| 78 |
262014_at |
AT1G35660
|
unknown |
0.703 |
0.025 |
| 79 |
258664_at |
AT3G08700
|
UBC12, ubiquitin-conjugating enzyme 12 |
0.695 |
0.040 |
| 80 |
262641_at |
AT1G62730
|
[Terpenoid synthases superfamily protein] |
0.695 |
0.016 |
| 81 |
263881_at |
AT2G21820
|
unknown |
0.692 |
-0.035 |
| 82 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
0.692 |
0.006 |
| 83 |
263732_at |
AT1G59980
|
ARL2, ARG1-like 2, ATDJC39, GPS4, gravity persistence signal 4 |
0.686 |
0.011 |
| 84 |
246612_at |
AT5G35320
|
unknown |
0.678 |
0.029 |
| 85 |
244923_s_at |
ATMG01020
|
ORF153B |
0.663 |
0.004 |
| 86 |
245313_at |
AT4G15420
|
[Ubiquitin fusion degradation UFD1 family protein] |
0.660 |
0.024 |
| 87 |
248426_at |
AT5G51740
|
[Peptidase family M48 family protein] |
0.660 |
0.017 |
| 88 |
264968_at |
AT1G67360
|
[Rubber elongation factor protein (REF)] |
0.657 |
-0.024 |
| 89 |
248774_at |
AT5G47830
|
unknown |
0.640 |
0.025 |
| 90 |
246981_at |
AT5G04840
|
[bZIP protein] |
0.636 |
-0.001 |
| 91 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
-0.710 |
0.110 |
| 92 |
259689_x_at |
AT1G63130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.680 |
0.019 |
| 93 |
254718_at |
AT4G13580
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.659 |
-0.003 |
| 94 |
263005_at |
AT1G54540
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.654 |
-0.036 |
| 95 |
253662_at |
AT4G30080
|
ARF16, auxin response factor 16 |
-0.640 |
-0.003 |
| 96 |
262105_at |
AT1G02810
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.620 |
0.046 |
| 97 |
253660_at |
AT4G30140
|
CDEF1, CUTICLE DESTRUCTING FACTOR 1 |
-0.583 |
-0.025 |
| 98 |
267645_at |
AT2G32860
|
BGLU33, beta glucosidase 33 |
-0.542 |
-0.013 |
| 99 |
260941_at |
AT1G44970
|
[Peroxidase superfamily protein] |
-0.511 |
0.070 |
| 100 |
245412_at |
AT4G17280
|
[Auxin-responsive family protein] |
-0.494 |
0.010 |
| 101 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
-0.470 |
-0.087 |
| 102 |
266222_at |
AT2G28780
|
unknown |
-0.444 |
0.073 |
| 103 |
259143_at |
AT3G10190
|
[Calcium-binding EF-hand family protein] |
-0.435 |
0.080 |
| 104 |
257690_at |
AT3G12830
|
SAUR72, SMALL AUXIN UPREGULATED 72 |
-0.432 |
0.042 |
| 105 |
266322_at |
AT2G46690
|
SAUR32, SMALL AUXIN UPREGULATED RNA 32 |
-0.427 |
0.014 |
| 106 |
262666_at |
AT1G14080
|
ATFUT6, FUT6, fucosyltransferase 6 |
-0.402 |
0.118 |
| 107 |
253065_at |
AT4G37740
|
AtGRF2, growth-regulating factor 2, GRF2, growth-regulating factor 2 |
-0.396 |
-0.070 |
| 108 |
255266_at |
AT4G05200
|
CRK25, cysteine-rich RLK (RECEPTOR-like protein kinase) 25 |
-0.392 |
0.052 |
| 109 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
-0.384 |
0.090 |
| 110 |
261700_at |
AT1G32690
|
unknown |
-0.384 |
0.012 |
| 111 |
261500_at |
AT1G28400
|
unknown |
-0.382 |
-0.029 |
| 112 |
256570_at |
AT3G19540
|
unknown |
-0.370 |
-0.014 |
| 113 |
249340_at |
AT5G41140
|
[Myosin heavy chain-related protein] |
-0.361 |
-0.044 |
| 114 |
263437_at |
AT2G28670
|
ESB1, ENHANCED SUBERIN 1 |
-0.358 |
-0.049 |
| 115 |
248056_at |
AT5G55500
|
ATXYLT, ARABIDOPSIS THALIANA BETA-1,2-XYLOSYLTRANSFERASE, XYLT, beta-1,2-xylosyltransferase |
-0.354 |
-0.004 |
| 116 |
266920_at |
AT2G45750
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.344 |
-0.032 |
| 117 |
252511_at |
AT3G46280
|
[protein kinase-related] |
-0.336 |
0.048 |
| 118 |
260582_at |
AT2G47200
|
unknown |
-0.330 |
0.038 |
| 119 |
264057_at |
AT2G28550
|
RAP2.7, related to AP2.7, TOE1, TARGET OF EARLY ACTIVATION TAGGED (EAT) 1 |
-0.326 |
-0.068 |
| 120 |
264078_at |
AT2G28470
|
BGAL8, beta-galactosidase 8 |
-0.323 |
-0.064 |
| 121 |
249970_at |
AT5G19100
|
[Eukaryotic aspartyl protease family protein] |
-0.318 |
-0.024 |
| 122 |
262040_at |
AT1G80080
|
AtRLP17, Receptor Like Protein 17, TMM, TOO MANY MOUTHS |
-0.316 |
-0.099 |
| 123 |
250438_at |
AT5G10580
|
unknown |
-0.316 |
0.025 |
| 124 |
245816_at |
AT1G26210
|
ATSOFL1, SOB FIVE-LIKE 1, SOFL1, SOB five-like 1 |
-0.302 |
-0.077 |
| 125 |
252692_at |
AT3G43960
|
[Cysteine proteinases superfamily protein] |
-0.291 |
-0.071 |
| 126 |
262788_at |
AT1G10690
|
unknown |
-0.283 |
-0.054 |
| 127 |
264685_at |
AT1G65610
|
ATGH9A2, ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9A2, KOR2, KORRIGAN 2 |
-0.281 |
0.045 |
| 128 |
248772_at |
AT5G47800
|
[Phototropic-responsive NPH3 family protein] |
-0.271 |
0.029 |
| 129 |
251672_at |
AT3G57230
|
AGL16, AGAMOUS-like 16 |
-0.269 |
-0.031 |
| 130 |
252272_at |
AT3G49670
|
BAM2, BARELY ANY MERISTEM 2 |
-0.269 |
-0.021 |
| 131 |
255348_at |
AT4G03820
|
unknown |
-0.267 |
-0.039 |
| 132 |
256488_at |
AT1G31470
|
NFD4, NUCLEAR FUSION DEFECTIVE 4 |
-0.267 |
-0.017 |
| 133 |
259812_at |
AT1G49840
|
unknown |
-0.265 |
-0.022 |
| 134 |
263406_at |
AT2G04160
|
AIR3, AUXIN-INDUCED IN ROOT CULTURES 3 |
-0.264 |
-0.008 |
| 135 |
256293_at |
AT1G69440
|
AGO7, ARGONAUTE7, ZIP, ZIPPY |
-0.263 |
0.032 |
| 136 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
-0.259 |
-0.146 |
| 137 |
248778_at |
AT5G47940
|
unknown |
-0.258 |
-0.046 |
| 138 |
246249_at |
AT4G36680
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.257 |
0.060 |
| 139 |
260076_at |
AT1G73630
|
[EF hand calcium-binding protein family] |
-0.255 |
0.030 |
| 140 |
264264_at |
AT1G09250
|
AIF4, ATBS1 Interacting Factor 4 |
-0.251 |
-0.045 |
| 141 |
250122_at |
AT5G16520
|
unknown |
-0.249 |
0.002 |
| 142 |
256781_at |
AT3G13650
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.248 |
-0.006 |
| 143 |
252209_at |
AT3G50400
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.248 |
-0.038 |
| 144 |
249916_at |
AT5G22880
|
H2B, HISTONE H2B, HTB2, histone B2 |
-0.248 |
-0.041 |
| 145 |
251015_at |
AT5G02480
|
[HSP20-like chaperones superfamily protein] |
-0.248 |
-0.012 |
| 146 |
245794_at |
AT1G32170
|
XTH30, xyloglucan endotransglucosylase/hydrolase 30, XTR4, xyloglucan endotransglycosylase 4 |
-0.246 |
0.006 |
| 147 |
250710_at |
AT5G06100
|
ATMYB33, MYB33, myb domain protein 33 |
-0.242 |
-0.047 |
| 148 |
249910_at |
AT5G22630
|
ADT5, arogenate dehydratase 5 |
-0.241 |
-0.004 |
| 149 |
255740_at |
AT1G25390
|
[Protein kinase superfamily protein] |
-0.241 |
0.030 |
| 150 |
248366_at |
AT5G52510
|
SCL8, SCARECROW-like 8 |
-0.239 |
-0.008 |
| 151 |
256626_at |
AT3G20015
|
[Eukaryotic aspartyl protease family protein] |
-0.238 |
-0.076 |
| 152 |
267534_at |
AT2G41900
|
OXS2, OXIDATIVE STRESS 2, TZF7, tandem zinc finger protein 7 |
-0.237 |
0.002 |
| 153 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
-0.233 |
0.049 |
| 154 |
252084_at |
AT3G51970
|
ASAT1, acyl-CoA sterol acyl transferase 1, ATASAT1, ATSAT1, ARABIDOPSIS THALIANA STEROL O-ACYLTRANSFERASE 1 |
-0.233 |
0.023 |
| 155 |
249020_at |
AT5G44800
|
CHR4, chromatin remodeling 4, PKR1, PICKLE RELATED 1 |
-0.232 |
0.009 |
| 156 |
256237_at |
AT3G12610
|
DRT100, DNA-DAMAGE REPAIR/TOLERATION 100 |
-0.230 |
-0.066 |
| 157 |
263242_at |
AT2G31400
|
GUN1, genomes uncoupled 1 |
-0.229 |
0.002 |
| 158 |
264828_at |
AT1G03380
|
ATATG18G, homolog of yeast autophagy 18 (ATG18) G, ATG18G, homolog of yeast autophagy 18 (ATG18) G |
-0.228 |
-0.026 |
| 159 |
252057_at |
AT3G52480
|
unknown |
-0.227 |
-0.024 |
| 160 |
250745_at |
AT5G05850
|
PIRL1, plant intracellular ras group-related LRR 1 |
-0.225 |
-0.061 |
| 161 |
258298_at |
AT3G23300
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.224 |
0.020 |
| 162 |
256622_at |
AT3G28920
|
AtHB34, homeobox protein 34, HB34, homeobox protein 34, ZHD9, ZINC FINGER HOMEODOMAIN 9 |
-0.223 |
0.026 |
| 163 |
249019_at |
AT5G44780
|
MORF4, multiple organellar RNA editing factor 4 |
-0.223 |
0.022 |
| 164 |
267110_at |
AT2G14800
|
unknown |
-0.219 |
0.026 |
| 165 |
251840_at |
AT3G54960
|
ATPDI1, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 1, ATPDIL1-3, PDI-like 1-3, PDI1, PROTEIN DISULFIDE ISOMERASE 1, PDIL1-3, PDI-like 1-3 |
-0.219 |
0.028 |
| 166 |
250682_x_at |
AT5G06630
|
[proline-rich extensin-like family protein] |
-0.217 |
-0.053 |
| 167 |
247922_at |
AT5G57500
|
[Galactosyltransferase family protein] |
-0.216 |
-0.025 |
| 168 |
245666_at |
AT1G28280
|
[VQ motif-containing protein] |
-0.216 |
0.061 |
| 169 |
262388_at |
AT1G49320
|
unknown |
-0.214 |
0.073 |
| 170 |
255467_at |
AT4G03010
|
[RNI-like superfamily protein] |
-0.214 |
-0.050 |
| 171 |
264689_at |
AT1G09900
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
-0.214 |
0.024 |
| 172 |
247856_at |
AT5G58300
|
[Leucine-rich repeat protein kinase family protein] |
-0.213 |
-0.009 |
| 173 |
267613_at |
AT2G26700
|
PID2, PINOID2 |
-0.210 |
0.012 |
| 174 |
253073_at |
AT4G37410
|
CYP81F4, cytochrome P450, family 81, subfamily F, polypeptide 4 |
-0.208 |
0.059 |
| 175 |
254260_at |
AT4G23440
|
[Disease resistance protein (TIR-NBS class)] |
-0.207 |
-0.017 |
| 176 |
253650_at |
AT4G30020
|
[PA-domain containing subtilase family protein] |
-0.207 |
-0.085 |
| 177 |
261050_at |
AT1G01260
|
JAM2, Jasmonate Associated MYC2 LIKE 2 |
-0.206 |
0.052 |
| 178 |
253531_at |
AT4G31540
|
ATEXO70G1, exocyst subunit exo70 family protein G1, EXO70G1, exocyst subunit exo70 family protein G1 |
-0.204 |
0.026 |
| 179 |
266995_at |
AT2G34500
|
CYP710A1, cytochrome P450, family 710, subfamily A, polypeptide 1 |
-0.204 |
0.082 |