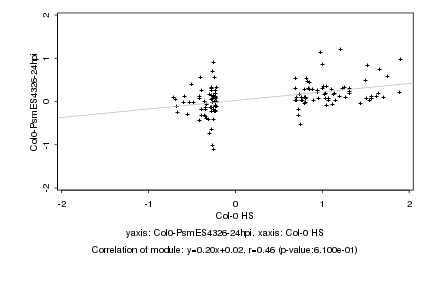
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Col-0 HS |
Log2 signal ratio
Col0-PsmES4326-24hpi |
| 1 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
1.896 |
0.992 |
| 2 |
262148_at |
AT1G52560
|
[HSP20-like chaperones superfamily protein] |
1.885 |
0.225 |
| 3 |
260978_at |
AT1G53540
|
[HSP20-like chaperones superfamily protein] |
1.740 |
0.586 |
| 4 |
262307_at |
AT1G71000
|
[Chaperone DnaJ-domain superfamily protein] |
1.700 |
0.116 |
| 5 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
1.655 |
0.763 |
| 6 |
253884_at |
AT4G27670
|
HSP21, heat shock protein 21 |
1.636 |
0.190 |
| 7 |
254059_at |
AT4G25200
|
ATHSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6, HSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6 |
1.615 |
0.125 |
| 8 |
251166_at |
AT3G63350
|
AT-HSFA7B, HSFA7B, HEAT SHOCK TRANSCRIPTION FACTOR A7B |
1.565 |
0.073 |
| 9 |
258695_at |
AT3G09640
|
APX1B, ASCORBATE PEROXIDASE 1B, APX2, ascorbate peroxidase 2 |
1.563 |
0.122 |
| 10 |
255811_at |
AT4G10250
|
ATHSP22.0 |
1.541 |
0.046 |
| 11 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
1.515 |
0.840 |
| 12 |
261838_at |
AT1G16030
|
Hsp70b, heat shock protein 70B |
1.506 |
0.088 |
| 13 |
249575_at |
AT5G37670
|
[HSP20-like chaperones superfamily protein] |
1.493 |
0.489 |
| 14 |
247691_at |
AT5G59720
|
HSP18.2, heat shock protein 18.2 |
1.430 |
-0.037 |
| 15 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
1.311 |
0.324 |
| 16 |
257323_at |
ATMG01200
|
ORF294 |
1.309 |
0.246 |
| 17 |
259913_at |
AT1G72660
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
1.304 |
0.190 |
| 18 |
248215_at |
AT5G53680
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
1.265 |
0.113 |
| 19 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
1.250 |
0.340 |
| 20 |
244902_at |
ATMG00650
|
NAD4L, NADH dehydrogenase subunit 4L |
1.223 |
0.317 |
| 21 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
1.201 |
1.223 |
| 22 |
258827_at |
AT3G07150
|
unknown |
1.188 |
0.132 |
| 23 |
244931_at |
ATMG00630
|
ORF110B |
1.143 |
0.040 |
| 24 |
245243_at |
AT1G44414
|
unknown |
1.133 |
0.206 |
| 25 |
257337_at |
ATMG00060
|
NAD5, NADH DEHYDROGENASE SUBUNIT 5, NAD5.3, NADH DEHYDROGENASE SUBUNIT 5.3, NAD5C, NADH dehydrogenase subunit 5C |
1.127 |
0.169 |
| 26 |
265675_at |
AT2G32120
|
HSP70T-2, heat-shock protein 70T-2 |
1.116 |
-0.059 |
| 27 |
254062_at |
AT4G25380
|
AtSAP10, Arabidopsis thaliana stress-associated protein 10, SAP10, stress-associated protein 10 |
1.095 |
0.296 |
| 28 |
254674_at |
AT4G18450
|
[Integrase-type DNA-binding superfamily protein] |
1.065 |
0.089 |
| 29 |
263164_at |
AT1G03070
|
AtLFG4, LFG4, LIFEGUARD 4 |
1.060 |
0.030 |
| 30 |
246944_at |
AT5G25450
|
[Cytochrome bd ubiquinol oxidase, 14kDa subunit] |
1.043 |
0.364 |
| 31 |
246733_at |
AT5G27660
|
DEG14, degradation of periplasmic proteins 14 |
1.042 |
-0.079 |
| 32 |
257321_at |
ATMG01130
|
ORF106F |
1.029 |
0.207 |
| 33 |
265242_at |
AT2G07705
|
unknown |
1.026 |
0.092 |
| 34 |
244943_at |
ATMG00070
|
NAD9, NADH dehydrogenase subunit 9 |
1.016 |
0.167 |
| 35 |
244901_at |
ATMG00640
|
ORF25 |
1.004 |
0.361 |
| 36 |
255651_at |
AT4G00940
|
AtDOF4.1, DOF4.1, DNA binding with one finger 4.1, ITD1, INTERCELLULAR TRAFFICKING DOF 1 |
1.000 |
0.312 |
| 37 |
254809_at |
AT4G12410
|
SAUR35, SMALL AUXIN UPREGULATED RNA 35 |
0.994 |
0.865 |
| 38 |
267026_at |
AT2G38340
|
DREB19, dehydration response element-binding protein 19 |
0.972 |
1.142 |
| 39 |
250036_at |
AT5G18340
|
[ARM repeat superfamily protein] |
0.947 |
0.084 |
| 40 |
263320_at |
AT2G47180
|
AtGolS1, galactinol synthase 1, GolS1, galactinol synthase 1 |
0.942 |
0.214 |
| 41 |
244950_at |
ATMG00160
|
COX2, cytochrome oxidase 2 |
0.936 |
0.274 |
| 42 |
244906_at |
ATMG00690
|
ORF240A |
0.890 |
0.037 |
| 43 |
262641_at |
AT1G62730
|
[Terpenoid synthases superfamily protein] |
0.884 |
0.285 |
| 44 |
250826_at |
AT5G05220
|
unknown |
0.850 |
0.444 |
| 45 |
253293_at |
AT4G33905
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
0.843 |
0.284 |
| 46 |
258406_at |
AT3G17611
|
ATRBL10, RHOMBOID-like protein 10, ATRBL14, RHOMBOID-like protein 14, RBL10, RHOMBOID-like protein 10, RBL14, RHOMBOID-like protein 14 |
0.834 |
0.318 |
| 47 |
253999_at |
AT4G26200
|
ACCS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ACS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ATACS7 |
0.826 |
0.466 |
| 48 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
0.807 |
0.539 |
| 49 |
250162_at |
AT5G15250
|
ATFTSH6, FTSH6, FTSH protease 6 |
0.805 |
0.098 |
| 50 |
251724_at |
AT3G56250
|
unknown |
0.804 |
-0.012 |
| 51 |
258336_at |
AT3G16050
|
A37, ATPDX1.2, ARABIDOPSIS THALIANA PYRIDOXINE BIOSYNTHESIS 1.2, PDX1.2, pyridoxine biosynthesis 1.2 |
0.804 |
0.116 |
| 52 |
253689_at |
AT4G29770
|
unknown |
0.799 |
0.082 |
| 53 |
256363_at |
AT1G66510
|
[AAR2 protein family] |
0.792 |
-0.026 |
| 54 |
244921_s_at |
ATMG01000
|
ORF114 |
0.792 |
0.101 |
| 55 |
246612_at |
AT5G35320
|
unknown |
0.791 |
0.280 |
| 56 |
257322_at |
ATMG01180
|
ORF111B |
0.771 |
0.037 |
| 57 |
254574_at |
AT4G19430
|
unknown |
0.754 |
0.106 |
| 58 |
267190_at |
AT2G44170
|
ATNMT2, ARABIDOPSIS N-MYRISTOYLTRANSFERASE 2, NMT2, N-myristoyltransferase 2 |
0.751 |
0.044 |
| 59 |
253949_at |
AT4G26780
|
AR192, MGE2, mitochondrial GrpE 2 |
0.739 |
-0.518 |
| 60 |
257874_at |
AT3G17110
|
[pseudogene] |
0.736 |
0.166 |
| 61 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.726 |
-0.308 |
| 62 |
265660_at |
AT2G25470
|
AtRLP21, receptor like protein 21, RLP21, receptor like protein 21 |
0.715 |
-0.165 |
| 63 |
257336_at |
ATMG01410
|
ORF204, open reading frame 204 |
0.700 |
0.109 |
| 64 |
244923_s_at |
ATMG01020
|
ORF153B |
0.697 |
0.035 |
| 65 |
261839_at |
AT1G16040
|
unknown |
0.688 |
0.313 |
| 66 |
257333_at |
ATMG01360
|
COX1, cytochrome oxidase |
0.688 |
0.028 |
| 67 |
253987_at |
AT4G26270
|
PFK3, phosphofructokinase 3 |
0.686 |
0.535 |
| 68 |
248178_at |
AT5G54370
|
[Late embryogenesis abundant (LEA) protein-related] |
-0.718 |
0.098 |
| 69 |
266131_at |
AT2G45160
|
ATHAM1, ARABIDOPSIS THALIANA HAIRY MERISTEM 1, HAM1, HAIRY MERISTEM 1, LOM1, LOST MERISTEMS 1 |
-0.694 |
0.051 |
| 70 |
267645_at |
AT2G32860
|
BGLU33, beta glucosidase 33 |
-0.681 |
-0.100 |
| 71 |
259689_x_at |
AT1G63130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.676 |
-0.247 |
| 72 |
247755_at |
AT5G59090
|
ATSBT4.12, subtilase 4.12, SBT4.12, subtilase 4.12 |
-0.606 |
-0.016 |
| 73 |
253662_at |
AT4G30080
|
ARF16, auxin response factor 16 |
-0.591 |
0.123 |
| 74 |
253794_at |
AT4G28720
|
YUC8, YUCCA 8 |
-0.559 |
-0.292 |
| 75 |
260618_at |
AT1G53230
|
TCP3, TEOSINTE BRANCHED 1, cycloidea and PCF transcription factor 3 |
-0.532 |
-0.013 |
| 76 |
262096_at |
AT1G56010
|
anac021, Arabidopsis NAC domain containing protein 21, ANAC022, Arabidopsis NAC domain containing protein 22, NAC1, NAC domain containing protein 1 |
-0.512 |
0.400 |
| 77 |
253078_at |
AT4G36180
|
[Leucine-rich receptor-like protein kinase family protein] |
-0.489 |
-0.001 |
| 78 |
252866_at |
AT4G39840
|
unknown |
-0.422 |
-0.427 |
| 79 |
266191_at |
AT2G39040
|
[Peroxidase superfamily protein] |
-0.419 |
0.086 |
| 80 |
245412_at |
AT4G17280
|
[Auxin-responsive family protein] |
-0.415 |
0.126 |
| 81 |
259282_at |
AT3G11430
|
ATGPAT5, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 5, GPAT5, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 5 |
-0.404 |
0.575 |
| 82 |
265831_at |
AT2G14460
|
unknown |
-0.398 |
-0.176 |
| 83 |
245088_at |
AT2G39850
|
[Subtilisin-like serine endopeptidase family protein] |
-0.397 |
-0.301 |
| 84 |
260565_at |
AT2G43800
|
[Actin-binding FH2 (formin homology 2) family protein] |
-0.397 |
-0.170 |
| 85 |
252100_at |
AT3G51110
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.393 |
0.270 |
| 86 |
261500_at |
AT1G28400
|
unknown |
-0.361 |
-0.314 |
| 87 |
262198_at |
AT1G53830
|
ATPME2, pectin methylesterase 2, PME2, pectin methylesterase 2 |
-0.359 |
0.001 |
| 88 |
267472_at |
AT2G02850
|
ARPN, plantacyanin |
-0.354 |
-0.119 |
| 89 |
247087_at |
AT5G66330
|
[Leucine-rich repeat (LRR) family protein] |
-0.352 |
-0.180 |
| 90 |
259579_at |
AT1G28010
|
ABCB14, ATP-binding cassette B14, ATABCB14, Arabidopsis thaliana ATP-binding cassette B14, MDR12, multi-drug resistance 12, PGP14, P-glycoprotein 14 |
-0.349 |
-0.336 |
| 91 |
258341_at |
AT3G22790
|
NET1A, Networked 1A |
-0.337 |
-0.054 |
| 92 |
251287_at |
AT3G61820
|
[Eukaryotic aspartyl protease family protein] |
-0.334 |
-0.389 |
| 93 |
262728_at |
AT1G75820
|
ATCLV1, CLV1, CLAVATA 1, FAS3, FASCIATA 3, FLO5, FLOWER DEVELOPMENT 5 |
-0.313 |
-0.401 |
| 94 |
248908_at |
AT5G45800
|
MEE62, maternal effect embryo arrest 62 |
-0.303 |
0.182 |
| 95 |
253255_at |
AT4G34760
|
SAUR50, SMALL AUXIN UPREGULATED RNA 50 |
-0.299 |
-0.729 |
| 96 |
246933_at |
AT5G25160
|
ZFP3, zinc finger protein 3 |
-0.294 |
-0.138 |
| 97 |
248201_at |
AT5G54180
|
PTAC15, plastid transcriptionally active 15 |
-0.287 |
-0.628 |
| 98 |
265102_at |
AT1G30870
|
[Peroxidase superfamily protein] |
-0.287 |
0.055 |
| 99 |
248006_at |
AT5G56250
|
HAP8, HAPLESS 8 |
-0.284 |
0.259 |
| 100 |
266223_at |
AT2G28790
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.280 |
-0.135 |
| 101 |
251895_at |
AT3G54420
|
ATCHITIV, CHITINASE CLASS IV, ATEP3, homolog of carrot EP3-3 chitinase, CHIV, EP3, homolog of carrot EP3-3 chitinase |
-0.279 |
0.312 |
| 102 |
248131_at |
AT5G54830
|
[DOMON domain-containing protein / dopamine beta-monooxygenase N-terminal domain-containing protein] |
-0.279 |
0.142 |
| 103 |
259661_at |
AT1G55265
|
unknown |
-0.278 |
0.340 |
| 104 |
256528_at |
AT1G66140
|
ZFP4, zinc finger protein 4 |
-0.277 |
-0.224 |
| 105 |
255281_at |
AT4G04970
|
ATGSL01, GLUCAN SYNTHASE LIKE 1, ATGSL1, GLUCAN SYNTHASE LIKE-1, GSL01, GSL1, glucan synthase-like 1 |
-0.274 |
-0.011 |
| 106 |
266322_at |
AT2G46690
|
SAUR32, SMALL AUXIN UPREGULATED RNA 32 |
-0.272 |
0.077 |
| 107 |
258912_at |
AT3G06460
|
[GNS1/SUR4 membrane protein family] |
-0.270 |
-0.015 |
| 108 |
253372_at |
AT4G33220
|
ATPME44, A. THALIANA PECTIN METHYLESTERASE 44, PME44, pectin methylesterase 44 |
-0.269 |
-1.003 |
| 109 |
264685_at |
AT1G65610
|
ATGH9A2, ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9A2, KOR2, KORRIGAN 2 |
-0.266 |
0.699 |
| 110 |
246478_at |
AT5G15980
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.261 |
-0.181 |
| 111 |
264828_at |
AT1G03380
|
ATATG18G, homolog of yeast autophagy 18 (ATG18) G, ATG18G, homolog of yeast autophagy 18 (ATG18) G |
-0.260 |
0.162 |
| 112 |
248945_at |
AT5G45510
|
[Leucine-rich repeat (LRR) family protein] |
-0.257 |
0.129 |
| 113 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
-0.256 |
0.907 |
| 114 |
256237_at |
AT3G12610
|
DRT100, DNA-DAMAGE REPAIR/TOLERATION 100 |
-0.254 |
-1.095 |
| 115 |
247383_at |
AT5G63410
|
[Leucine-rich repeat protein kinase family protein] |
-0.253 |
-0.408 |
| 116 |
261471_at |
AT1G14460
|
[AAA-type ATPase family protein] |
-0.250 |
-0.229 |
| 117 |
261099_at |
AT1G62980
|
ATEXP18, ATEXPA18, expansin A18, ATHEXP ALPHA 1.25, EXP18, EXPANSIN 18, EXPA18, expansin A18 |
-0.249 |
0.010 |
| 118 |
249910_at |
AT5G22630
|
ADT5, arogenate dehydratase 5 |
-0.247 |
0.556 |
| 119 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
-0.246 |
-0.213 |
| 120 |
257202_at |
AT3G23750
|
BARK1, BAK1-Associating Receptor-Like Kinase 1 |
-0.241 |
-0.189 |
| 121 |
248254_at |
AT5G53320
|
[Leucine-rich repeat protein kinase family protein] |
-0.239 |
-0.013 |
| 122 |
250399_at |
AT5G10750
|
unknown |
-0.239 |
0.108 |
| 123 |
249832_at |
AT5G23400
|
[Leucine-rich repeat (LRR) family protein] |
-0.237 |
-0.105 |
| 124 |
249969_at |
AT5G19090
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.237 |
-0.209 |
| 125 |
265573_at |
AT2G28200
|
[C2H2-type zinc finger family protein] |
-0.237 |
0.133 |
| 126 |
262750_at |
AT1G28710
|
[Nucleotide-diphospho-sugar transferase family protein] |
-0.236 |
-0.071 |
| 127 |
247554_at |
AT5G61010
|
ATEXO70E2, exocyst subunit exo70 family protein E2, EXO70E2, exocyst subunit exo70 family protein E2 |
-0.236 |
0.269 |
| 128 |
246920_at |
AT5G25090
|
AtENODL13, ENODL13, early nodulin-like protein 13 |
-0.234 |
0.076 |
| 129 |
260924_at |
AT1G21590
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.233 |
-0.102 |
| 130 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
-0.231 |
0.200 |
| 131 |
263218_at |
AT1G30570
|
HERK2, hercules receptor kinase 2 |
-0.229 |
-0.094 |
| 132 |
257093_at |
AT3G20570
|
AtENODL9, ENODL9, early nodulin-like protein 9 |
-0.227 |
0.017 |
| 133 |
253743_at |
AT4G28940
|
[Phosphorylase superfamily protein] |
-0.224 |
0.326 |