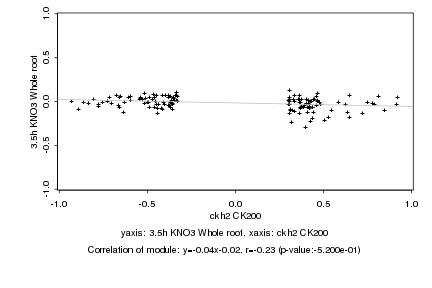
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ckh2 CK200 |
Log2 signal ratio
3.5h KNO3 Whole root |
| 1 |
260584_at |
AT2G43660
|
[Carbohydrate-binding X8 domain superfamily protein] |
0.917 |
0.049 |
| 2 |
257768_at |
AT3G23060
|
[RING/U-box superfamily protein] |
0.914 |
-0.024 |
| 3 |
249482_at |
AT5G38980
|
unknown |
0.844 |
-0.091 |
| 4 |
260706_at |
AT1G32350
|
AOX1D, alternative oxidase 1D |
0.808 |
0.061 |
| 5 |
253962_at |
AT4G26460
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.788 |
-0.033 |
| 6 |
253667_at |
AT4G30170
|
[Peroxidase family protein] |
0.774 |
-0.016 |
| 7 |
254371_at |
AT4G21760
|
BGLU47, beta-glucosidase 47 |
0.749 |
-0.006 |
| 8 |
260410_at |
AT1G69870
|
AtNPF2.13, NPF2.13, NRT1/ PTR family 2.13, NRT1.7, nitrate transporter 1.7 |
0.716 |
-0.126 |
| 9 |
248227_at |
AT5G53820
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.646 |
-0.172 |
| 10 |
261420_at |
AT1G07720
|
KCS3, 3-ketoacyl-CoA synthase 3 |
0.644 |
0.070 |
| 11 |
265117_at |
AT1G62500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.636 |
-0.125 |
| 12 |
255768_at |
AT1G16705
|
[p300/CBP acetyltransferase-related protein-related] |
0.623 |
-0.023 |
| 13 |
260541_at |
AT2G43530
|
[Scorpion toxin-like knottin superfamily protein] |
0.584 |
-0.005 |
| 14 |
262638_at |
AT1G06650
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.542 |
-0.098 |
| 15 |
251519_at |
AT3G59400
|
GUN4, GENOMES UNCOUPLED 4 |
0.523 |
-0.175 |
| 16 |
265111_at |
AT1G62510
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.502 |
-0.212 |
| 17 |
253871_at |
AT4G27440
|
PORB, protochlorophyllide oxidoreductase B |
0.482 |
-0.033 |
| 18 |
250006_at |
AT5G18660
|
PCB2, PALE-GREEN AND CHLOROPHYLL B REDUCED 2 |
0.477 |
-0.003 |
| 19 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
0.472 |
-0.010 |
| 20 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
0.464 |
0.013 |
| 21 |
246273_at |
AT4G36700
|
[RmlC-like cupins superfamily protein] |
0.461 |
0.009 |
| 22 |
253173_at |
AT4G35110
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.461 |
0.093 |
| 23 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
0.459 |
-0.043 |
| 24 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
0.457 |
0.062 |
| 25 |
259223_at |
AT3G03660
|
WOX11, WUSCHEL related homeobox 11 |
0.448 |
0.031 |
| 26 |
263182_at |
AT1G05575
|
unknown |
0.439 |
-0.117 |
| 27 |
246238_at |
AT4G36670
|
AtPLT6, AtPMT6, PLT6, polyol transporter 6, PMT6, polyol/monosaccharide transporter 6 |
0.439 |
0.034 |
| 28 |
252972_at |
AT4G38840
|
SAUR14, SMALL AUXIN UPREGULATED RNA 14 |
0.436 |
-0.191 |
| 29 |
256927_at |
AT3G22550
|
unknown |
0.435 |
-0.060 |
| 30 |
250435_at |
AT5G10380
|
ATRING1, RING1 |
0.434 |
-0.067 |
| 31 |
251036_at |
AT5G02160
|
unknown |
0.428 |
0.009 |
| 32 |
245765_at |
AT1G33600
|
[Leucine-rich repeat (LRR) family protein] |
0.425 |
0.011 |
| 33 |
262283_at |
AT1G68590
|
PSRP3/1, plastid-specific ribosomal protein 3/1 |
0.421 |
-0.219 |
| 34 |
255572_at |
AT4G01050
|
TROL, thylakoid rhodanese-like |
0.420 |
-0.077 |
| 35 |
267288_at |
AT2G23680
|
[Cold acclimation protein WCOR413 family] |
0.420 |
-0.032 |
| 36 |
265481_at |
AT2G15960
|
unknown |
0.419 |
-0.046 |
| 37 |
250669_at |
AT5G06870
|
ATPGIP2, ARABIDOPSIS POLYGALACTURONASE INHIBITING PROTEIN 2, PGIP2, polygalacturonase inhibiting protein 2 |
0.413 |
-0.016 |
| 38 |
263142_at |
AT1G65230
|
[Uncharacterized conserved protein (DUF2358)] |
0.411 |
0.021 |
| 39 |
261910_at |
AT1G80730
|
ATZFP1, ARABIDOPSIS THALIANA ZINC-FINGER PROTEIN 1, ZFP1, zinc-finger protein 1 |
0.408 |
-0.117 |
| 40 |
264839_at |
AT1G03630
|
POR C, protochlorophyllide oxidoreductase C, PORC |
0.407 |
-0.068 |
| 41 |
252136_at |
AT3G50770
|
CML41, calmodulin-like 41 |
0.402 |
0.025 |
| 42 |
266293_at |
AT2G29360
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.398 |
-0.016 |
| 43 |
247320_at |
AT5G64040
|
PSAN |
0.394 |
-0.287 |
| 44 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.389 |
-0.039 |
| 45 |
264301_at |
AT1G78780
|
[pathogenesis-related family protein] |
0.383 |
-0.065 |
| 46 |
267076_at |
AT2G41090
|
[Calcium-binding EF-hand family protein] |
0.371 |
0.032 |
| 47 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
0.370 |
-0.055 |
| 48 |
252116_at |
AT3G51510
|
unknown |
0.367 |
-0.062 |
| 49 |
264824_at |
AT1G03420
|
Sadhu4-2, sadhu non-coding retrotransposon 4-2 |
0.366 |
-0.070 |
| 50 |
267003_at |
AT2G34340
|
unknown |
0.365 |
-0.007 |
| 51 |
248348_at |
AT5G52190
|
[Sugar isomerase (SIS) family protein] |
0.362 |
-0.132 |
| 52 |
259834_at |
AT1G69570
|
[Dof-type zinc finger DNA-binding family protein] |
0.360 |
0.026 |
| 53 |
245143_at |
AT2G45450
|
ZPR1, LITTLE ZIPPER 1 |
0.359 |
0.077 |
| 54 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
0.359 |
-0.000 |
| 55 |
261216_at |
AT1G33030
|
[O-methyltransferase family protein] |
0.357 |
0.027 |
| 56 |
259552_at |
AT1G21320
|
[nucleotide binding] |
0.356 |
0.024 |
| 57 |
249694_at |
AT5G35790
|
G6PD1, glucose-6-phosphate dehydrogenase 1 |
0.335 |
0.009 |
| 58 |
247487_at |
AT5G62150
|
[peptidoglycan-binding LysM domain-containing protein] |
0.330 |
-0.105 |
| 59 |
261346_at |
AT1G79720
|
[Eukaryotic aspartyl protease family protein] |
0.330 |
0.079 |
| 60 |
259010_at |
AT3G07340
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.324 |
0.028 |
| 61 |
248007_at |
AT5G56260
|
[Ribonuclease E inhibitor RraA/Dimethylmenaquinone methyltransferase] |
0.321 |
-0.091 |
| 62 |
247899_at |
AT5G57345
|
unknown |
0.314 |
-0.230 |
| 63 |
266285_at |
AT2G29180
|
unknown |
0.312 |
-0.080 |
| 64 |
249147_at |
AT5G43330
|
c-NAD-MDH2, cytosolic-NAD-dependent malate dehydrogenase 2 |
0.309 |
-0.091 |
| 65 |
249255_at |
AT5G41610
|
ATCHX18, ARABIDOPSIS THALIANA CATION/H+ EXCHANGER 18, CHX18, cation/H+ exchanger 18 |
0.306 |
0.135 |
| 66 |
260696_at |
AT1G32520
|
unknown |
0.306 |
0.007 |
| 67 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.305 |
0.050 |
| 68 |
265287_at |
AT2G20260
|
PSAE-2, photosystem I subunit E-2 |
0.303 |
-0.133 |
| 69 |
258941_at |
AT3G09940
|
ATMDAR3, ARABIDOPSIS THALIANA MONODEHYDROASCORBATE REDUCTASE 3, MDAR2, MONODEHYDROASCORBATE REDUCTASE 2, MDAR3, MONODEHYDROASCORBATE REDUCTASE 3, MDHAR, monodehydroascorbate reductase |
0.302 |
0.034 |
| 70 |
251899_at |
AT3G54400
|
[Eukaryotic aspartyl protease family protein] |
0.302 |
-0.032 |
| 71 |
249784_at |
AT5G24280
|
GMI1, GAMMA-IRRADIATION AND MITOMYCIN C INDUCED 1 |
0.298 |
0.015 |
| 72 |
251983_at |
AT3G53210
|
UMAMIT6, Usually multiple acids move in and out Transporters 6 |
0.297 |
-0.024 |
| 73 |
263098_at |
AT2G16005
|
[MD-2-related lipid recognition domain-containing protein] |
-0.935 |
0.005 |
| 74 |
254907_at |
AT4G11190
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.894 |
-0.090 |
| 75 |
253084_at |
AT4G36260
|
SRS2, SHI RELATED SEQUENCE 2, STY2, STYLISH 2 |
-0.863 |
-0.006 |
| 76 |
252363_at |
AT3G48460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.835 |
-0.021 |
| 77 |
253483_at |
AT4G31910
|
BAT1, BR-related AcylTransferase1, PIZ, PIZZA |
-0.809 |
0.029 |
| 78 |
263979_at |
AT2G42840
|
PDF1, protodermal factor 1 |
-0.782 |
-0.047 |
| 79 |
262819_at |
AT1G11600
|
CYP77B1, cytochrome P450, family 77, subfamily B, polypeptide 1 |
-0.781 |
-0.033 |
| 80 |
255111_at |
AT4G08780
|
[Peroxidase superfamily protein] |
-0.755 |
-0.000 |
| 81 |
254034_at |
AT4G25960
|
ABCB2, ATP-binding cassette B2, PGP2, P-glycoprotein 2 |
-0.731 |
0.009 |
| 82 |
264137_at |
AT1G78960
|
ATLUP2, lupeol synthase 2, LUP2, lupeol synthase 2 |
-0.716 |
0.054 |
| 83 |
253754_at |
AT4G29020
|
[glycine-rich protein] |
-0.706 |
-0.019 |
| 84 |
252943_at |
AT4G39330
|
ATCAD9, CAD9, cinnamyl alcohol dehydrogenase 9 |
-0.679 |
0.070 |
| 85 |
256750_at |
AT3G27150
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.665 |
-0.042 |
| 86 |
253054_at |
AT4G37580
|
COP3, CONSTITUTIVE PHOTOMORPHOGENIC 3, HLS1, HOOKLESS 1, UNS2, UNUSUAL SUGAR RESPONSE 2 |
-0.664 |
-0.067 |
| 87 |
253684_at |
AT4G29690
|
[Alkaline-phosphatase-like family protein] |
-0.664 |
0.056 |
| 88 |
262659_at |
AT1G14240
|
[GDA1/CD39 nucleoside phosphatase family protein] |
-0.654 |
0.062 |
| 89 |
258675_at |
AT3G08770
|
LTP6, lipid transfer protein 6 |
-0.638 |
-0.119 |
| 90 |
259350_at |
AT3G05140
|
RBK2, ROP binding protein kinases 2 |
-0.633 |
-0.008 |
| 91 |
251160_at |
AT3G63240
|
[DNAse I-like superfamily protein] |
-0.609 |
0.054 |
| 92 |
258253_at |
AT3G26760
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.601 |
0.023 |
| 93 |
256030_at |
AT1G34110
|
[Leucine-rich receptor-like protein kinase family protein] |
-0.598 |
0.058 |
| 94 |
267377_at |
AT2G26250
|
FDH, FIDDLEHEAD, KCS10, 3-ketoacyl-CoA synthase 10 |
-0.547 |
0.027 |
| 95 |
246908_at |
AT5G25610
|
ATRD22, RD22, RESPONSIVE TO DESSICATION 22 |
-0.542 |
0.046 |
| 96 |
262376_at |
AT1G72970
|
EDA17, embryo sac development arrest 17, HTH, HOTHEAD |
-0.541 |
0.044 |
| 97 |
251668_at |
AT3G57010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.532 |
0.034 |
| 98 |
249567_at |
AT5G38020
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.522 |
0.098 |
| 99 |
248736_at |
AT5G48110
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.518 |
-0.013 |
| 100 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
-0.511 |
0.039 |
| 101 |
253011_at |
AT4G37890
|
EDA40, embryo sac development arrest 40 |
-0.500 |
-0.005 |
| 102 |
248732_at |
AT5G48070
|
ATXTH20, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 20, XTH20, xyloglucan endotransglucosylase/hydrolase 20 |
-0.497 |
-0.011 |
| 103 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
-0.491 |
-0.061 |
| 104 |
245140_at |
AT2G45420
|
LBD18, LOB domain-containing protein 18 |
-0.489 |
0.056 |
| 105 |
262414_at |
AT1G49430
|
LACS2, long-chain acyl-CoA synthetase 2, LRD2, LATERAL ROOT DEVELOPMENT 2 |
-0.474 |
0.033 |
| 106 |
245141_at |
AT2G45400
|
BEN1, BRI1-5 ENHANCED 1 |
-0.468 |
0.085 |
| 107 |
264470_at |
AT1G67110
|
CYP735A2, cytochrome P450, family 735, subfamily A, polypeptide 2 |
-0.463 |
-0.057 |
| 108 |
249557_at |
AT5G38280
|
PR5K, PR5-like receptor kinase |
-0.461 |
0.049 |
| 109 |
263909_at |
AT2G36490
|
AtROS1, DML1, demeter-like 1, ROS1, REPRESSOR OF SILENCING1 |
-0.454 |
0.010 |
| 110 |
261878_at |
AT1G50560
|
CYP705A25, cytochrome P450, family 705, subfamily A, polypeptide 25 |
-0.453 |
-0.025 |
| 111 |
247884_at |
AT5G57800
|
CER3, ECERIFERUM 3, FLP1, FACELESS POLLEN 1, WAX2, YRE |
-0.451 |
0.069 |
| 112 |
248070_at |
AT5G55660
|
[DEK domain-containing chromatin associated protein] |
-0.447 |
-0.132 |
| 113 |
262680_at |
AT1G75880
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.445 |
-0.078 |
| 114 |
250499_at |
AT5G09730
|
ATBX3, ATBXL3, BETA-XYLOSIDASE 3, BX3, BXL3, beta-xylosidase 3, XYL3 |
-0.437 |
-0.030 |
| 115 |
254392_at |
AT4G21600
|
ENDO5, endonuclease 5 |
-0.425 |
-0.073 |
| 116 |
261109_at |
AT1G75450
|
ATCKX5, ARABIDOPSIS THALIANA CYTOKININ OXIDASE 5, ATCKX6, CYTOKININ OXIDASE 6, CKX5, cytokinin oxidase 5 |
-0.418 |
0.070 |
| 117 |
252128_at |
AT3G50870
|
GATA18, GATA TRANSCRIPTION FACTOR 18, HAN, HANABA TANARU, MNP, MONOPOLE |
-0.417 |
-0.081 |
| 118 |
260943_at |
AT1G45145
|
ATH5, THIOREDOXIN H-TYPE 5, ATTRX5, thioredoxin H-type 5, LIV1, LOCUS OF INSENSITIVITY TO VICTORIN 1, TRX-h5, THIOREDOXIN H-TYPE 5, TRX5, thioredoxin H-type 5 |
-0.412 |
-0.008 |
| 119 |
251466_at |
AT3G59340
|
unknown |
-0.408 |
-0.027 |
| 120 |
266838_at |
AT2G25980
|
[Mannose-binding lectin superfamily protein] |
-0.400 |
0.070 |
| 121 |
266596_at |
AT2G46150
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.385 |
0.055 |
| 122 |
253406_at |
AT4G32890
|
GATA9, GATA transcription factor 9 |
-0.384 |
-0.034 |
| 123 |
264384_at |
AT2G25170
|
CHD3, CHR6, CKH2, CYTOKININ-HYPERSENSITIVE 2, EPP1, ENHANCED PHOTOMORPHOGENIC 1, GYM, GYMNOS, PKL, PICKLE, SSL2, SUPPRESSOR OF SLR 2 |
-0.383 |
0.076 |
| 124 |
257105_at |
AT3G15300
|
[VQ motif-containing protein] |
-0.379 |
-0.041 |
| 125 |
258021_at |
AT3G19380
|
PUB25, plant U-box 25 |
-0.372 |
-0.058 |
| 126 |
261315_at |
AT1G53170
|
ATERF-8, ATERF8, ETHYLENE RESPONSE ELEMENT BINDING FACTOR 4, ERF8, ethylene response factor 8 |
-0.372 |
-0.005 |
| 127 |
252321_at |
AT3G48510
|
unknown |
-0.369 |
0.058 |
| 128 |
247297_at |
AT5G64100
|
[Peroxidase superfamily protein] |
-0.367 |
0.022 |
| 129 |
253437_at |
AT4G32460
|
unknown |
-0.367 |
-0.006 |
| 130 |
266532_at |
AT2G16890
|
[UDP-Glycosyltransferase superfamily protein] |
-0.361 |
-0.082 |
| 131 |
263594_at |
AT2G01880
|
ATPAP7, PURPLE ACID PHOSPHATASE 7, PAP7, purple acid phosphatase 7 |
-0.360 |
-0.013 |
| 132 |
245382_at |
AT4G17800
|
AHL23, AT-hook motif nuclear localized protein 23 |
-0.358 |
-0.030 |
| 133 |
262538_at |
AT1G17140
|
ICR1, interactor of constitutive active rops 1, RIP1, ROP INTERACTIVE PARTNER 1 |
-0.357 |
0.044 |
| 134 |
254226_at |
AT4G23690
|
AtDIR6, Arabidopsis thaliana dirigent protein 6, DIR6, dirigent protein 6 |
-0.354 |
0.041 |
| 135 |
254122_at |
AT4G24510
|
CER2, ECERIFERUM 2, VC-2, VC2 |
-0.353 |
0.047 |
| 136 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.347 |
0.021 |
| 137 |
262971_at |
AT1G75640
|
[Leucine-rich receptor-like protein kinase family protein] |
-0.344 |
0.071 |
| 138 |
264318_at |
AT1G04220
|
KCS2, 3-ketoacyl-CoA synthase 2 |
-0.337 |
0.021 |
| 139 |
248584_at |
AT5G49960
|
unknown |
-0.337 |
0.076 |
| 140 |
257923_at |
AT3G23160
|
unknown |
-0.335 |
0.114 |
| 141 |
266737_at |
AT2G47140
|
AtSDR5, SDR5, short-chain dehydrogenase reductase 5 |
-0.334 |
0.008 |
| 142 |
258599_at |
AT3G04520
|
THA2, threonine aldolase 2 |
-0.333 |
0.007 |
| 143 |
259680_at |
AT1G77690
|
LAX3, like AUX1 3 |
-0.330 |
0.065 |