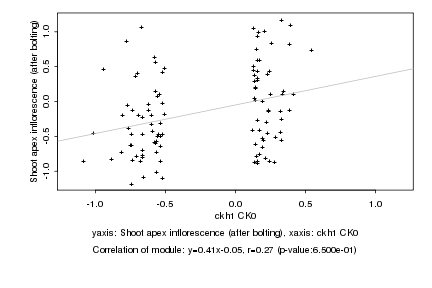
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ckh1 CK0 |
Log2 signal ratio
Shoot apex inflorescence (after bolting) |
| 1 |
259640_at |
AT1G52400
|
ATBG1, A. THALIANA BETA-GLUCOSIDASE 1, BGL1, BETA-GLUCOSIDASE HOMOLOG 1, BGLU18, beta glucosidase 18 |
0.540 |
0.742 |
| 2 |
250722_at |
AT5G06190
|
unknown |
0.411 |
0.110 |
| 3 |
265530_at |
AT2G06050
|
AtOPR3, DDE1, DELAYED DEHISCENCE 1, OPR3, oxophytodienoate-reductase 3 |
0.390 |
1.103 |
| 4 |
248104_at |
AT5G55250
|
AtIAMT1, IAMT1, IAA carboxylmethyltransferase 1 |
0.386 |
0.829 |
| 5 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
0.384 |
-0.119 |
| 6 |
263709_at |
AT1G09310
|
unknown |
0.341 |
0.145 |
| 7 |
258560_at |
AT3G06020
|
FAF4, FANTASTIC FOUR 4 |
0.330 |
0.107 |
| 8 |
245692_at |
AT5G04150
|
BHLH101 |
0.327 |
-0.556 |
| 9 |
267376_at |
AT2G26330
|
ER, ERECTA, QRP1, QUANTITATIVE RESISTANCE TO PLECTOSPHAERELLA 1 |
0.327 |
1.167 |
| 10 |
264626_at |
AT1G65620
|
AS2, ASYMMETRIC LEAVES 2 |
0.324 |
-0.255 |
| 11 |
254256_at |
AT4G23180
|
CRK10, cysteine-rich RLK (RECEPTOR-like protein kinase) 10, RLK4 |
0.321 |
-0.131 |
| 12 |
262680_at |
AT1G75880
|
[SGNH hydrolase-type esterase superfamily protein] |
0.316 |
-0.432 |
| 13 |
250670_at |
AT5G06860
|
ATPGIP1, POLYGALACTURONASE INHIBITING PROTEIN 1, PGIP1, polygalacturonase inhibiting protein 1 |
0.280 |
-0.510 |
| 14 |
247515_at |
AT5G61740
|
ABCA10, ATP-binding cassette A10, ATATH14, ARABIDOPSIS THALIANA ABC2 HOMOLOG 14, ATH14, ABC2 homolog 14 |
0.276 |
-0.862 |
| 15 |
257929_at |
AT3G16980
|
NRPB9A, NRPD9A, NRPE9A |
0.251 |
0.837 |
| 16 |
265276_at |
AT2G28400
|
unknown |
0.244 |
0.107 |
| 17 |
258608_at |
AT3G03020
|
unknown |
0.241 |
0.438 |
| 18 |
246943_at |
AT5G25440
|
[Protein kinase superfamily protein] |
0.239 |
-0.849 |
| 19 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.235 |
-0.131 |
| 20 |
265205_at |
AT2G36660
|
PAB7, poly(A) binding protein 7 |
0.233 |
-0.127 |
| 21 |
260101_at |
AT1G73260
|
ATKTI1, ARABIDOPSIS THALIANA KUNITZ TRYPSIN INHIBITOR 1, KTI1, kunitz trypsin inhibitor 1 |
0.228 |
0.396 |
| 22 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
0.224 |
-0.455 |
| 23 |
266739_at |
AT2G46730
|
[pseudogene] |
0.216 |
-0.299 |
| 24 |
264751_at |
AT1G23020
|
ATFRO3, FERRIC REDUCTION OXIDASE 3, FRO3, ferric reduction oxidase 3 |
0.214 |
-0.810 |
| 25 |
261220_at |
AT1G19970
|
[ER lumen protein retaining receptor family protein] |
0.201 |
1.016 |
| 26 |
266749_at |
AT2G47060
|
PTI1-4, Pto-interacting 1-4 |
0.197 |
-0.546 |
| 27 |
261535_at |
AT1G01725
|
unknown |
0.190 |
-0.646 |
| 28 |
260401_at |
AT1G69840
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
0.188 |
0.001 |
| 29 |
248740_at |
AT5G48130
|
[Phototropic-responsive NPH3 family protein] |
0.188 |
-0.519 |
| 30 |
254524_at |
AT4G20000
|
[VQ motif-containing protein] |
0.171 |
-0.404 |
| 31 |
258008_at |
AT3G19430
|
[late embryogenesis abundant protein-related / LEA protein-related] |
0.167 |
-0.755 |
| 32 |
258510_at |
AT3G06600
|
unknown |
0.165 |
0.602 |
| 33 |
246908_at |
AT5G25610
|
ATRD22, RD22, RESPONSIVE TO DESSICATION 22 |
0.163 |
1.000 |
| 34 |
256837_at |
AT3G22900
|
NRPD7 |
0.156 |
0.444 |
| 35 |
263808_at |
AT2G04340
|
unknown |
0.155 |
-0.883 |
| 36 |
252408_at |
AT3G47600
|
ATMYB94, myb domain protein 94, ATMYBCP70, MYB94, myb domain protein 94 |
0.155 |
-0.267 |
| 37 |
252245_at |
AT3G49710
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.153 |
0.338 |
| 38 |
264318_at |
AT1G04220
|
KCS2, 3-ketoacyl-CoA synthase 2 |
0.153 |
0.312 |
| 39 |
257168_at |
AT3G24430
|
HCF101, HIGH-CHLOROPHYLL-FLUORESCENCE 101 |
0.153 |
0.596 |
| 40 |
250762_at |
AT5G05990
|
[Mitochondrial glycoprotein family protein] |
0.152 |
0.933 |
| 41 |
263158_at |
AT1G54160
|
NF-YA5, nuclear factor Y, subunit A5, NFYA5, NUCLEAR FACTOR Y A5 |
0.151 |
-0.847 |
| 42 |
262046_at |
AT1G79960
|
ATOFP14, OFP14, ovate family protein 14 |
0.150 |
-0.783 |
| 43 |
245768_at |
AT1G33590
|
[Leucine-rich repeat (LRR) family protein] |
0.147 |
0.756 |
| 44 |
253882_at |
AT4G27650
|
PEL1, PELOTA |
0.140 |
0.211 |
| 45 |
262338_at |
AT1G64185
|
[Lactoylglutathione lyase / glyoxalase I family protein] |
0.138 |
0.017 |
| 46 |
265894_at |
AT2G15050
|
LTP, lipid transfer protein, LTP7, lipid transfer protein 7 |
0.137 |
-0.604 |
| 47 |
250382_at |
AT5G11580
|
[Regulator of chromosome condensation (RCC1) family protein] |
0.137 |
0.196 |
| 48 |
246132_at |
AT5G20850
|
ATRAD51, RAD51 |
0.136 |
0.291 |
| 49 |
253780_at |
AT4G28400
|
[Protein phosphatase 2C family protein] |
0.135 |
0.386 |
| 50 |
257373_at |
AT2G43140
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.134 |
-0.872 |
| 51 |
254287_at |
AT4G22960
|
unknown |
0.132 |
0.053 |
| 52 |
266601_at |
AT2G46060
|
[transmembrane protein-related] |
0.123 |
0.453 |
| 53 |
261681_at |
AT1G47340
|
[F-box and associated interaction domains-containing protein] |
0.122 |
1.059 |
| 54 |
246762_at |
AT5G27620
|
CYCH;1, cyclin H;1 |
0.122 |
0.515 |
| 55 |
256646_at |
AT3G13590
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.121 |
-0.410 |
| 56 |
259328_at |
AT3G16440
|
ATMLP-300B, myrosinase-binding protein-like protein-300B, MEE36, maternal effect embryo arrest 36, MLP-300B, myrosinase-binding protein-like protein-300B |
-1.087 |
-0.850 |
| 57 |
267119_at |
AT2G32610
|
ATCSLB01, cellulose synthase-like B1, ATCSLB1, CELLULOSE SYNTHASE LIKE B1, CSLB01, CELLULOSE SYNTHASE LIKE B1, CSLB01, cellulose synthase-like B1 |
-1.017 |
-0.447 |
| 58 |
254907_at |
AT4G11190
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.947 |
0.468 |
| 59 |
250059_at |
AT5G17820
|
[Peroxidase superfamily protein] |
-0.891 |
-0.824 |
| 60 |
260492_at |
AT2G41850
|
ADPG2, ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE 2, PGAZAT, polygalacturonase abscission zone A. thaliana |
-0.815 |
-0.731 |
| 61 |
258293_at |
AT3G23430
|
ATPHO1, ARABIDOPSIS PHOSPHATE 1, PHO1, phosphate 1 |
-0.808 |
-0.198 |
| 62 |
264987_at |
AT1G27030
|
unknown |
-0.780 |
0.865 |
| 63 |
258769_at |
AT3G10870
|
ATMES17, ARABIDOPSIS THALIANA METHYL ESTERASE 17, MES17, methyl esterase 17 |
-0.777 |
-0.047 |
| 64 |
250100_at |
AT5G16570
|
GLN1;4, glutamine synthetase 1;4 |
-0.770 |
-0.385 |
| 65 |
247696_at |
AT5G59780
|
ATMYB59, MYB DOMAIN PROTEIN 59, ATMYB59-1, ATMYB59-2, ATMYB59-3, MYB59, myb domain protein 59 |
-0.751 |
-0.619 |
| 66 |
262260_at |
AT1G70850
|
MLP34, MLP-like protein 34 |
-0.748 |
-1.176 |
| 67 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
-0.748 |
-0.626 |
| 68 |
265119_at |
AT1G62570
|
FMO GS-OX4, flavin-monooxygenase glucosinolate S-oxygenase 4 |
-0.746 |
-0.469 |
| 69 |
251595_at |
AT3G57620
|
[glyoxal oxidase-related protein] |
-0.738 |
-0.837 |
| 70 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
-0.737 |
-0.119 |
| 71 |
245386_at |
AT4G14010
|
RALFL32, ralf-like 32 |
-0.715 |
0.362 |
| 72 |
248736_at |
AT5G48110
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.710 |
-0.777 |
| 73 |
256815_at |
AT3G21380
|
[Mannose-binding lectin superfamily protein] |
-0.704 |
0.402 |
| 74 |
263282_at |
AT2G14095
|
unknown |
-0.698 |
-0.200 |
| 75 |
263098_at |
AT2G16005
|
[MD-2-related lipid recognition domain-containing protein] |
-0.680 |
-0.859 |
| 76 |
254225_at |
AT4G23670
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.673 |
1.065 |
| 77 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
-0.670 |
-0.801 |
| 78 |
254674_at |
AT4G18450
|
[Integrase-type DNA-binding superfamily protein] |
-0.669 |
-0.219 |
| 79 |
249097_at |
AT5G43520
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.668 |
-0.768 |
| 80 |
260335_at |
AT1G74000
|
SS3, strictosidine synthase 3 |
-0.667 |
-0.466 |
| 81 |
254013_at |
AT4G26050
|
PIRL8, plant intracellular ras group-related LRR 8 |
-0.666 |
-0.700 |
| 82 |
260890_at |
AT1G29090
|
[Cysteine proteinases superfamily protein] |
-0.659 |
-1.078 |
| 83 |
260559_at |
AT2G43860
|
[Pectin lyase-like superfamily protein] |
-0.628 |
-0.117 |
| 84 |
262454_at |
AT1G11190
|
BFN1, bifunctional nuclease i, ENDO1, ENDONUCLEASE 1 |
-0.624 |
-0.031 |
| 85 |
256515_at |
AT1G66020
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.605 |
-0.328 |
| 86 |
266866_at |
AT2G29940
|
ABCG31, ATP-binding cassette G31, ATPDR3, PLEIOTROPIC DRUG RESISTANCE 3, PDR3, pleiotropic drug resistance 3 |
-0.605 |
-0.190 |
| 87 |
261425_at |
AT1G18880
|
AtNPF2.9, NPF2.9, NRT1/ PTR family 2.9, NRT1.9, nitrate transporter 1.9 |
-0.597 |
-0.426 |
| 88 |
253660_at |
AT4G30140
|
CDEF1, CUTICLE DESTRUCTING FACTOR 1 |
-0.583 |
-0.577 |
| 89 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
-0.580 |
0.644 |
| 90 |
264102_at |
AT1G79270
|
ECT8, evolutionarily conserved C-terminal region 8 |
-0.578 |
-0.593 |
| 91 |
267038_at |
AT2G38480
|
[Uncharacterised protein family (UPF0497)] |
-0.576 |
0.561 |
| 92 |
249567_at |
AT5G38020
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.575 |
0.157 |
| 93 |
250142_at |
AT5G14650
|
[Pectin lyase-like superfamily protein] |
-0.568 |
-0.562 |
| 94 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.566 |
-0.729 |
| 95 |
252004_at |
AT3G52780
|
ATPAP20, PAP20 |
-0.566 |
-1.006 |
| 96 |
263689_at |
AT1G26820
|
RNS3, ribonuclease 3 |
-0.563 |
0.078 |
| 97 |
253332_at |
AT4G33420
|
[Peroxidase superfamily protein] |
-0.560 |
-0.497 |
| 98 |
267548_at |
AT2G32660
|
AtRLP22, receptor like protein 22, RLP22, receptor like protein 22 |
-0.555 |
-0.459 |
| 99 |
262637_at |
AT1G06640
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.548 |
0.106 |
| 100 |
264782_at |
AT1G08810
|
AtMYB60, myb domain protein 60, MYB60, myb domain protein 60 |
-0.542 |
-0.857 |
| 101 |
257309_at |
AT3G28150
|
AXY4L, ALTERED XYLOGLUCAN 4-LIKE, TBL22, TRICHOME BIREFRINGENCE-LIKE 22 |
-0.542 |
-0.308 |
| 102 |
257761_at |
AT3G23090
|
WDL3 |
-0.539 |
-0.489 |
| 103 |
252006_at |
AT3G52820
|
ATPAP22, PURPLE ACID PHOSPHATASE 22, PAP22, purple acid phosphatase 22 |
-0.536 |
-0.642 |
| 104 |
262972_at |
AT1G75620
|
[glyoxal oxidase-related protein] |
-0.528 |
-0.470 |
| 105 |
256011_at |
AT1G19230
|
[Riboflavin synthase-like superfamily protein] |
-0.527 |
-1.095 |
| 106 |
266322_at |
AT2G46690
|
SAUR32, SMALL AUXIN UPREGULATED RNA 32 |
-0.524 |
-0.028 |
| 107 |
253073_at |
AT4G37410
|
CYP81F4, cytochrome P450, family 81, subfamily F, polypeptide 4 |
-0.522 |
0.427 |
| 108 |
250045_at |
AT5G17700
|
[MATE efflux family protein] |
-0.511 |
0.481 |
| 109 |
267127_at |
AT2G23610
|
ATMES3, ARABIDOPSIS THALIANA METHYL ESTERASE 3, MES3, methyl esterase 3 |
-0.510 |
-0.186 |