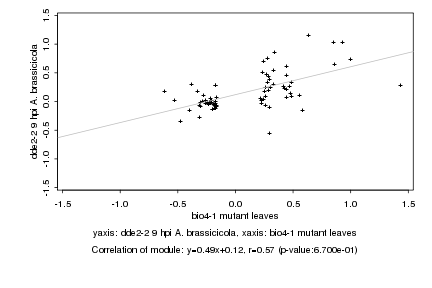
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
bio4-1 mutant leaves |
Log2 signal ratio
dde2-2 9 hpi A. brassicicola |
| 1 |
254387_at |
AT4G21850
|
ATMSRB9, methionine sulfoxide reductase B9, MSRB9, methionine sulfoxide reductase B9 |
1.429 |
0.284 |
| 2 |
254832_at |
AT4G12490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.995 |
0.739 |
| 3 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
0.925 |
1.039 |
| 4 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
0.854 |
0.655 |
| 5 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.843 |
1.036 |
| 6 |
264616_at |
AT2G17740
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.633 |
1.164 |
| 7 |
266246_at |
AT2G27690
|
CYP94C1, cytochrome P450, family 94, subfamily C, polypeptide 1 |
0.577 |
-0.146 |
| 8 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
0.550 |
0.109 |
| 9 |
254093_at |
AT4G25110
|
AtMC2, metacaspase 2, AtMCP1c, metacaspase 1c, MC2, metacaspase 2, MCP1c, metacaspase 1c |
0.482 |
0.343 |
| 10 |
264680_at |
AT1G65510
|
unknown |
0.481 |
0.101 |
| 11 |
253485_at |
AT4G31800
|
ATWRKY18, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 18, WRKY18, WRKY DNA-binding protein 18 |
0.475 |
0.152 |
| 12 |
248551_at |
AT5G50200
|
ATNRT3.1, NRT3.1, NITRATE TRANSPORTER 3.1, WR3, WOUND-RESPONSIVE 3 |
0.467 |
0.263 |
| 13 |
246133_at |
AT5G20960
|
AAO1, aldehyde oxidase 1, AO1, aldehyde oxidase 1, AOalpha, aldehyde oxidase alpha, AT-AO1, ARABIDOPSIS THALIANA ALDEHYDE OXIDASE 1, ATAO, AtAO1, Arabidopsis thaliana aldehyde oxidase 1 |
0.441 |
0.464 |
| 14 |
245317_at |
AT4G15610
|
[Uncharacterised protein family (UPF0497)] |
0.439 |
0.618 |
| 15 |
249377_at |
AT5G40690
|
unknown |
0.436 |
0.085 |
| 16 |
255596_at |
AT4G01720
|
AtWRKY47, WRKY47 |
0.436 |
0.221 |
| 17 |
262542_at |
AT1G34180
|
anac016, NAC domain containing protein 16, NAC016, NAC domain containing protein 16 |
0.424 |
0.231 |
| 18 |
249889_at |
AT5G22540
|
unknown |
0.410 |
0.278 |
| 19 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
0.338 |
0.858 |
| 20 |
264746_at |
AT1G62300
|
ATWRKY6, WRKY6 |
0.330 |
0.557 |
| 21 |
257950_at |
AT3G21780
|
UGT71B6, UDP-glucosyl transferase 71B6 |
0.329 |
0.310 |
| 22 |
265028_at |
AT1G24530
|
[Transducin/WD40 repeat-like superfamily protein] |
0.304 |
0.251 |
| 23 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
0.294 |
0.389 |
| 24 |
255879_at |
AT1G67000
|
[Protein kinase superfamily protein] |
0.289 |
-0.100 |
| 25 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.288 |
-0.552 |
| 26 |
248037_at |
AT5G55930
|
ATOPT1, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 1, OPT1, oligopeptide transporter 1 |
0.282 |
0.207 |
| 27 |
255479_at |
AT4G02380
|
AtLEA5, Arabidopsis thaliana late embryogenensis abundant like 5, SAG21, senescence-associated gene 21 |
0.280 |
0.442 |
| 28 |
254408_at |
AT4G21390
|
B120 |
0.277 |
0.765 |
| 29 |
262324_at |
AT1G64170
|
ATCHX16, cation/H+ exchanger 16, CHX16, cation/H+ exchanger 16 |
0.271 |
0.344 |
| 30 |
263411_at |
AT2G28710
|
[C2H2-type zinc finger family protein] |
0.269 |
0.479 |
| 31 |
255999_at |
AT1G29860
|
ATWRKY71, WRKY DNA-BINDING PROTEIN 71, WRKY71, WRKY DNA-binding protein 71 |
0.260 |
0.245 |
| 32 |
259320_at |
AT3G01080
|
ATWRKY58, WRKY DNA-BINDING PROTEIN 58, WRKY58, WRKY DNA-binding protein 58 |
0.257 |
-0.053 |
| 33 |
251398_at |
AT3G60580
|
[C2H2-like zinc finger protein] |
0.253 |
0.094 |
| 34 |
262444_at |
AT1G47480
|
[alpha/beta-Hydrolases superfamily protein] |
0.248 |
0.188 |
| 35 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.241 |
0.044 |
| 36 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
0.239 |
0.702 |
| 37 |
259841_at |
AT1G52200
|
[PLAC8 family protein] |
0.234 |
0.522 |
| 38 |
247850_at |
AT5G58150
|
[Leucine-rich repeat protein kinase family protein] |
0.224 |
0.021 |
| 39 |
248092_at |
AT5G55170
|
ATSUMO3, SUM3, SMALL UBIQUITIN-LIKE MODIFIER 3, SUMO 3, SMALL UBIQUITIN-LIKE MODIFIER 3, SUMO3, small ubiquitin-like modifier 3 |
0.222 |
-0.020 |
| 40 |
260531_at |
AT2G47240
|
CER8, ECERIFERUM 8, LACS1, LONG-CHAIN ACYL-COA SYNTHASE 1 |
0.212 |
0.064 |
| 41 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.620 |
0.188 |
| 42 |
264741_at |
AT1G62290
|
[Saposin-like aspartyl protease family protein] |
-0.537 |
0.028 |
| 43 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
-0.484 |
-0.342 |
| 44 |
254716_at |
AT4G13560
|
UNE15, unfertilized embryo sac 15 |
-0.400 |
-0.155 |
| 45 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
-0.382 |
0.302 |
| 46 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
-0.335 |
0.176 |
| 47 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.319 |
-0.059 |
| 48 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
-0.318 |
-0.263 |
| 49 |
250937_at |
AT5G03230
|
unknown |
-0.311 |
-0.007 |
| 50 |
250528_at |
AT5G08600
|
[U3 ribonucleoprotein (Utp) family protein] |
-0.304 |
-0.072 |
| 51 |
260773_at |
AT1G78440
|
ATGA2OX1, Arabidopsis thaliana gibberellin 2-oxidase 1, GA2OX1, gibberellin 2-oxidase 1 |
-0.290 |
0.014 |
| 52 |
257650_at |
AT3G16800
|
[Protein phosphatase 2C family protein] |
-0.281 |
0.108 |
| 53 |
248867_at |
AT5G46830
|
ATNIG1, NACL-INDUCIBLE GENE 1, NIG1, NACL-inducible gene 1 |
-0.267 |
-0.022 |
| 54 |
262212_at |
AT1G74890
|
ARR15, response regulator 15 |
-0.267 |
0.020 |
| 55 |
264130_at |
AT1G79160
|
unknown |
-0.247 |
-0.001 |
| 56 |
251668_at |
AT3G57010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.245 |
-0.026 |
| 57 |
250372_at |
AT5G11460
|
unknown |
-0.237 |
-0.044 |
| 58 |
261610_at |
AT1G49560
|
[Homeodomain-like superfamily protein] |
-0.228 |
-0.012 |
| 59 |
251169_at |
AT3G63210
|
MARD1, MEDIATOR OF ABA-REGULATED DORMANCY 1 |
-0.223 |
-0.022 |
| 60 |
250327_at |
AT5G12050
|
unknown |
-0.222 |
0.058 |
| 61 |
262164_at |
AT1G78070
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.219 |
-0.030 |
| 62 |
246783_at |
AT5G27360
|
SFP2 |
-0.215 |
-0.012 |
| 63 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
-0.210 |
0.020 |
| 64 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.201 |
-0.133 |
| 65 |
265740_at |
AT2G01150
|
RHA2B, RING-H2 finger protein 2B |
-0.200 |
-0.134 |
| 66 |
251058_at |
AT5G01790
|
unknown |
-0.193 |
-0.042 |
| 67 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
-0.189 |
-0.047 |
| 68 |
264252_at |
AT1G09180
|
ATSAR1, SECRETION-ASSOCIATED RAS 1, ATSARA1A, secretion-associated RAS super family 1, SARA1A, secretion-associated RAS super family 1 |
-0.185 |
-0.052 |
| 69 |
250233_at |
AT5G13460
|
IQD11, IQ-domain 11 |
-0.184 |
-0.110 |
| 70 |
266824_at |
AT2G22800
|
HAT9 |
-0.181 |
0.283 |
| 71 |
264314_at |
AT1G70420
|
unknown |
-0.181 |
0.281 |
| 72 |
254175_at |
AT4G24050
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.180 |
-0.024 |
| 73 |
249843_at |
AT5G23570
|
ATSGS3, SUPPRESSOR OF GENE SILENCING 3, SGS3, SUPPRESSOR OF GENE SILENCING 3 |
-0.180 |
-0.119 |
| 74 |
252594_at |
AT3G45680
|
[Major facilitator superfamily protein] |
-0.179 |
-0.080 |
| 75 |
252983_at |
AT4G37980
|
ATCAD7, CAD7, CINNAMYL-ALCOHOL DEHYDROGENASE 7, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-1, elicitor-activated gene 3-1 |
-0.178 |
0.004 |
| 76 |
255768_at |
AT1G16705
|
[p300/CBP acetyltransferase-related protein-related] |
-0.174 |
-0.067 |
| 77 |
245075_at |
AT2G23180
|
CYP96A1, cytochrome P450, family 96, subfamily A, polypeptide 1 |
-0.170 |
-0.079 |
| 78 |
260639_at |
AT1G53180
|
unknown |
-0.169 |
0.072 |
| 79 |
253525_at |
AT4G31330
|
unknown |
-0.167 |
-0.058 |