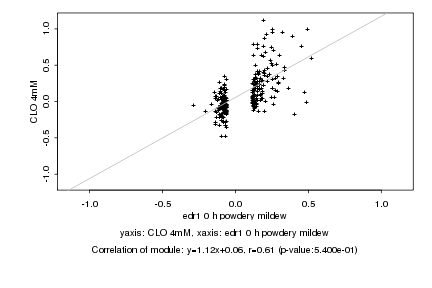
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
edr1 0 h powdery mildew |
Log2 signal ratio
CLO 4mM |
| 1 |
264648_at |
AT1G09080
|
BIP3, binding protein 3 |
0.515 |
0.594 |
| 2 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.489 |
1.004 |
| 3 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
0.482 |
-0.002 |
| 4 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
0.469 |
0.135 |
| 5 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
0.449 |
0.763 |
| 6 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
0.401 |
-0.178 |
| 7 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
0.386 |
0.903 |
| 8 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
0.357 |
0.187 |
| 9 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
0.334 |
0.436 |
| 10 |
259385_at |
AT1G13470
|
unknown |
0.333 |
0.480 |
| 11 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
0.324 |
0.319 |
| 12 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
0.318 |
0.960 |
| 13 |
253181_at |
AT4G35180
|
LHT7, LYS/HIS transporter 7 |
0.298 |
0.644 |
| 14 |
247685_at |
AT5G59680
|
[Leucine-rich repeat protein kinase family protein] |
0.292 |
0.266 |
| 15 |
260904_at |
AT1G02450
|
NIMIN-1, NIMIN1, NIM1-interacting 1 |
0.291 |
0.255 |
| 16 |
253044_at |
AT4G37290
|
unknown |
0.283 |
0.346 |
| 17 |
245393_at |
AT4G16260
|
[Glycosyl hydrolase superfamily protein] |
0.283 |
0.140 |
| 18 |
254252_at |
AT4G23310
|
CRK23, cysteine-rich RLK (RECEPTOR-like protein kinase) 23 |
0.280 |
0.518 |
| 19 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
0.273 |
0.159 |
| 20 |
263852_at |
AT2G04450
|
ATNUDT6, nudix hydrolase homolog 6, ATNUDX6, Arabidopsis thaliana nucleoside diphosphate linked to some moiety X 6, NUDT6, nudix hydrolase homolog 6, NUDX6, nucleoside diphosphates linked to some moiety X 6 |
0.268 |
0.321 |
| 21 |
267385_at |
AT2G44380
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.264 |
0.063 |
| 22 |
257100_at |
AT3G25010
|
AtRLP41, receptor like protein 41, RLP41, receptor like protein 41 |
0.257 |
0.711 |
| 23 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
0.255 |
-0.032 |
| 24 |
247314_at |
AT5G64000
|
ATSAL2, SAL2 |
0.253 |
0.500 |
| 25 |
258983_at |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
0.252 |
0.193 |
| 26 |
256596_at |
AT3G28540
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.252 |
0.305 |
| 27 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
0.250 |
0.188 |
| 28 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
0.249 |
0.963 |
| 29 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
0.248 |
0.993 |
| 30 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
0.245 |
0.056 |
| 31 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
0.243 |
0.757 |
| 32 |
255406_at |
AT4G03450
|
[Ankyrin repeat family protein] |
0.241 |
0.533 |
| 33 |
265161_at |
AT1G30900
|
BP80-3;3, binding protein of 80 kDa 3;3, VSR3;3, VACUOLAR SORTING RECEPTOR 3;3, VSR6, VACUOLAR SORTING RECEPTOR 6 |
0.238 |
0.565 |
| 34 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
0.232 |
0.381 |
| 35 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
0.218 |
0.354 |
| 36 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
0.214 |
0.283 |
| 37 |
254253_at |
AT4G23320
|
CRK24, cysteine-rich RLK (RECEPTOR-like protein kinase) 24 |
0.214 |
0.457 |
| 38 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.209 |
0.936 |
| 39 |
265327_at |
AT2G18210
|
unknown |
0.201 |
0.003 |
| 40 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
0.200 |
0.688 |
| 41 |
263536_at |
AT2G25000
|
ATWRKY60, WRKY60, WRKY DNA-binding protein 60 |
0.198 |
0.387 |
| 42 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.197 |
0.371 |
| 43 |
265471_at |
AT2G37130
|
[Peroxidase superfamily protein] |
0.194 |
0.434 |
| 44 |
256442_at |
AT3G10930
|
unknown |
0.193 |
0.876 |
| 45 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
0.193 |
-0.129 |
| 46 |
259040_at |
AT3G09270
|
ATGSTU8, glutathione S-transferase TAU 8, GSTU8, glutathione S-transferase TAU 8 |
0.192 |
0.770 |
| 47 |
254387_at |
AT4G21850
|
ATMSRB9, methionine sulfoxide reductase B9, MSRB9, methionine sulfoxide reductase B9 |
0.191 |
0.406 |
| 48 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
0.190 |
0.233 |
| 49 |
246993_at |
AT5G67450
|
AZF1, zinc-finger protein 1, ZF1, zinc-finger protein 1 |
0.188 |
0.620 |
| 50 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
0.187 |
1.129 |
| 51 |
248330_at |
AT5G52810
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.187 |
0.232 |
| 52 |
260126_at |
AT1G36370
|
SHM7, serine hydroxymethyltransferase 7 |
0.183 |
0.046 |
| 53 |
248169_at |
AT5G54610
|
ANK, ankyrin, BDA1, bian da 2 (becoming big in Chinese) |
0.182 |
0.117 |
| 54 |
264958_at |
AT1G76960
|
unknown |
0.182 |
0.138 |
| 55 |
265189_at |
AT1G23840
|
unknown |
0.181 |
0.294 |
| 56 |
254343_at |
AT4G21990
|
APR3, APS reductase 3, ATAPR3, PRH-26, PRH26, PAPS REDUCTASE HOMOLOG 26 |
0.177 |
0.048 |
| 57 |
252549_at |
AT3G45860
|
CRK4, cysteine-rich RLK (RECEPTOR-like protein kinase) 4 |
0.177 |
0.190 |
| 58 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.175 |
0.643 |
| 59 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.172 |
0.276 |
| 60 |
247800_at |
AT5G58570
|
unknown |
0.172 |
0.279 |
| 61 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
0.170 |
0.319 |
| 62 |
259384_at |
AT3G16450
|
JAL33, Jacalin-related lectin 33 |
0.168 |
0.017 |
| 63 |
261177_at |
AT1G04770
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.167 |
0.027 |
| 64 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.165 |
0.098 |
| 65 |
265722_at |
AT2G40100
|
LHCB4.3, light harvesting complex photosystem II |
0.165 |
0.093 |
| 66 |
259992_at |
AT1G67970
|
AT-HSFA8, HSFA8, heat shock transcription factor A8 |
0.164 |
0.168 |
| 67 |
254500_at |
AT4G20110
|
BP80-3;1, binding protein of 80 kDa 3;1, VSR3;1, VACUOLAR SORTING RECEPTOR 3;1, VSR7, VACUOLAR SORTING RECEPTOR 7 |
0.163 |
0.241 |
| 68 |
245982_at |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
0.163 |
0.279 |
| 69 |
263073_at |
AT2G17500
|
[Auxin efflux carrier family protein] |
0.160 |
0.281 |
| 70 |
250090_at |
AT5G17330
|
GAD, glutamate decarboxylase, GAD1, GLUTAMATE DECARBOXYLASE 1 |
0.160 |
0.422 |
| 71 |
255941_at |
AT1G20350
|
ATTIM17-1, translocase inner membrane subunit 17-1, TIM17-1, translocase inner membrane subunit 17-1 |
0.160 |
0.190 |
| 72 |
247714_at |
AT5G59340
|
WOX2, WUSCHEL related homeobox 2 |
0.156 |
-0.124 |
| 73 |
254839_at |
AT4G12400
|
Hop3, Hop3 |
0.153 |
0.381 |
| 74 |
254453_at |
AT4G21120
|
AAT1, amino acid transporter 1, CAT1, CATIONIC AMINO ACID TRANSPORTER 1 |
0.152 |
0.386 |
| 75 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.150 |
0.416 |
| 76 |
258791_at |
AT3G04720
|
AtPR4, HEL, HEVEIN-LIKE, PR-4, PR4, pathogenesis-related 4 |
0.149 |
0.145 |
| 77 |
262605_at |
AT1G15170
|
[MATE efflux family protein] |
0.149 |
0.090 |
| 78 |
246366_at |
AT1G51850
|
[Leucine-rich repeat protein kinase family protein] |
0.149 |
0.285 |
| 79 |
266988_at |
AT2G39310
|
JAL22, jacalin-related lectin 22 |
0.148 |
0.026 |
| 80 |
245611_at |
AT4G14390
|
[Ankyrin repeat family protein] |
0.147 |
0.211 |
| 81 |
260978_at |
AT1G53540
|
[HSP20-like chaperones superfamily protein] |
0.147 |
0.743 |
| 82 |
254430_at |
AT4G20820
|
[FAD-binding Berberine family protein] |
0.146 |
0.183 |
| 83 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
0.145 |
0.786 |
| 84 |
253957_at |
AT4G26320
|
AGP13, arabinogalactan protein 13 |
0.144 |
-0.062 |
| 85 |
256969_at |
AT3G21080
|
[ABC transporter-related] |
0.141 |
0.049 |
| 86 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
0.141 |
-0.019 |
| 87 |
250435_at |
AT5G10380
|
ATRING1, RING1 |
0.141 |
0.251 |
| 88 |
264261_at |
AT1G09240
|
ATNAS3, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 3, NAS3, nicotianamine synthase 3 |
0.140 |
0.260 |
| 89 |
250446_at |
AT5G10770
|
[Eukaryotic aspartyl protease family protein] |
0.139 |
-0.036 |
| 90 |
255430_at |
AT4G03320
|
AtTic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV, Tic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV |
0.139 |
0.319 |
| 91 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
0.136 |
0.190 |
| 92 |
256833_at |
AT3G22910
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.136 |
0.380 |
| 93 |
259211_at |
AT3G09020
|
[alpha 1,4-glycosyltransferase family protein] |
0.135 |
0.336 |
| 94 |
265132_at |
AT1G23830
|
unknown |
0.135 |
0.221 |
| 95 |
251975_at |
AT3G53230
|
AtCDC48B, cell division cycle 48B |
0.135 |
0.265 |
| 96 |
252862_at |
AT4G39830
|
[Cupredoxin superfamily protein] |
0.134 |
0.501 |
| 97 |
260648_at |
AT1G08050
|
[Zinc finger (C3HC4-type RING finger) family protein] |
0.133 |
0.317 |
| 98 |
261167_at |
AT1G04980
|
ATPDI10, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 10, ATPDIL2-2, PDI-like 2-2, PDI10, PROTEIN DISULFIDE ISOMERASE, PDIL2-2, PDI-like 2-2 |
0.130 |
0.107 |
| 99 |
254630_at |
AT4G18360
|
GOX3, glycolate oxidase 3 |
0.130 |
0.188 |
| 100 |
262912_at |
AT1G59740
|
AtNPF4.3, NPF4.3, NRT1/ PTR family 4.3 |
0.129 |
-0.013 |
| 101 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
0.127 |
-0.028 |
| 102 |
253907_at |
AT4G27250
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.126 |
-0.073 |
| 103 |
256162_at |
AT1G55390
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.126 |
0.002 |
| 104 |
255284_at |
AT4G04610
|
APR, APR1, APS reductase 1, ATAPR1, PRH19, PAPS REDUCTASE HOMOLOG 19 |
0.125 |
-0.060 |
| 105 |
259852_at |
AT1G72280
|
AERO1, endoplasmic reticulum oxidoreductins 1, ERO1, endoplasmic reticulum oxidoreductins 1 |
0.125 |
0.286 |
| 106 |
256641_at |
AT3G32270
|
unknown |
0.125 |
-0.087 |
| 107 |
245769_at |
AT1G30220
|
ATINT2, ARABIDOPSIS THALIANA INOSITOL TRANSPORTER 2, INT2, inositol transporter 2 |
0.125 |
0.154 |
| 108 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
0.124 |
-0.057 |
| 109 |
259766_at |
AT1G64360
|
unknown |
0.124 |
0.081 |
| 110 |
258932_at |
AT3G10150
|
ATPAP16, PAP16, purple acid phosphatase 16 |
0.124 |
0.018 |
| 111 |
266778_at |
AT2G29090
|
CYP707A2, cytochrome P450, family 707, subfamily A, polypeptide 2 |
0.123 |
-0.045 |
| 112 |
258063_at |
AT3G14620
|
CYP72A8, cytochrome P450, family 72, subfamily A, polypeptide 8 |
0.123 |
0.182 |
| 113 |
257480_at |
AT1G15640
|
unknown |
0.123 |
0.038 |
| 114 |
265624_at |
AT2G27250
|
AtCLV3, CLV3, CLAVATA3 |
0.123 |
-0.004 |
| 115 |
258338_at |
AT3G16150
|
ASPGB1, asparaginase B1 |
0.123 |
-0.002 |
| 116 |
251419_at |
AT3G60470
|
unknown |
0.121 |
0.088 |
| 117 |
255932_at |
AT1G12720
|
[Mutator-like transposase family, has a 1.2e-40 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
0.121 |
-0.055 |
| 118 |
257592_at |
AT3G24982
|
ATRLP40, receptor like protein 40, RLP40, receptor like protein 40 |
0.120 |
0.797 |
| 119 |
254247_at |
AT4G23260
|
CRK18, cysteine-rich RLK (RECEPTOR-like protein kinase) 18 |
0.120 |
0.237 |
| 120 |
251842_at |
AT3G54580
|
[Proline-rich extensin-like family protein] |
0.120 |
0.014 |
| 121 |
248134_at |
AT5G54860
|
[Major facilitator superfamily protein] |
0.119 |
0.094 |
| 122 |
259173_at |
AT3G03640
|
BGLU25, beta glucosidase 25, GLUC |
0.119 |
0.034 |
| 123 |
266711_at |
AT2G46740
|
AtGulLO5, GulLO5, L -gulono-1,4-lactone ( L -GulL) oxidase 5 |
0.119 |
-0.043 |
| 124 |
259925_at |
AT1G75040
|
PR-5, PR5, pathogenesis-related gene 5 |
0.118 |
0.634 |
| 125 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
0.118 |
0.313 |
| 126 |
256762_at |
AT3G25655
|
IDL1, inflorescence deficient in abscission (IDA)-like 1 |
0.118 |
0.027 |
| 127 |
255302_at |
AT4G04830
|
ATMSRB5, methionine sulfoxide reductase B5, MSRB5, methionine sulfoxide reductase B5 |
0.117 |
0.007 |
| 128 |
250682_x_at |
AT5G06630
|
[proline-rich extensin-like family protein] |
0.117 |
-0.110 |
| 129 |
264832_at |
AT1G03660
|
[Ankyrin-repeat containing protein] |
0.117 |
0.156 |
| 130 |
252921_at |
AT4G39030
|
EDS5, ENHANCED DISEASE SUSCEPTIBILITY 5, SCORD3, susceptible to coronatine-deficient Pst DC3000 3, SID1, SALICYLIC ACID INDUCTION DEFICIENT 1 |
0.116 |
0.137 |
| 131 |
262607_at |
AT1G13990
|
unknown |
0.115 |
0.064 |
| 132 |
256444_at |
AT3G11060
|
unknown |
0.114 |
0.051 |
| 133 |
246228_at |
AT4G36430
|
[Peroxidase superfamily protein] |
0.114 |
0.027 |
| 134 |
267496_at |
AT2G30550
|
DALL3, DAD1-Like Lipase 3 |
0.113 |
0.161 |
| 135 |
256619_at |
AT3G24460
|
[Serinc-domain containing serine and sphingolipid biosynthesis protein] |
0.112 |
-0.009 |
| 136 |
251093_at |
AT5G01360
|
TBL3, TRICHOME BIREFRINGENCE-LIKE 3 |
0.112 |
-0.017 |
| 137 |
264734_at |
AT1G62280
|
SLAH1, SLAC1 homologue 1 |
0.112 |
0.070 |
| 138 |
246597_at |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
0.111 |
0.250 |
| 139 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
-0.294 |
-0.050 |
| 140 |
250561_at |
AT5G08030
|
AtGDPD6, GDPD6, glycerophosphodiester phosphodiesterase 6 |
-0.207 |
-0.135 |
| 141 |
259001_at |
AT3G01960
|
unknown |
-0.170 |
-0.039 |
| 142 |
257207_at |
AT3G14900
|
EMB3120, EMBRYO DEFECTIVE 3120 |
-0.147 |
0.135 |
| 143 |
261037_at |
AT1G17420
|
ATLOX3, Arabidopsis thaliana lipoxygenase 3, LOX3, lipoxygenase 3 |
-0.142 |
-0.126 |
| 144 |
256248_at |
AT3G66652
|
[fip1 motif-containing protein] |
-0.141 |
0.070 |
| 145 |
262891_at |
AT1G79460
|
ATKS, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE, ATKS1, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE 1, GA2, GA REQUIRING 2, KS, KS1, ENT-KAURENE SYNTHASE 1 |
-0.140 |
-0.277 |
| 146 |
256702_at |
AT3G30380
|
[alpha/beta-Hydrolases superfamily protein] |
-0.139 |
0.052 |
| 147 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
-0.137 |
-0.307 |
| 148 |
251494_at |
AT3G59350
|
[Protein kinase superfamily protein] |
-0.135 |
0.070 |
| 149 |
256302_at |
AT1G69526
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.134 |
-0.219 |
| 150 |
264549_at |
AT1G09440
|
[Protein kinase superfamily protein] |
-0.133 |
-0.143 |
| 151 |
261983_at |
AT1G33770
|
[Protein kinase superfamily protein] |
-0.128 |
0.015 |
| 152 |
245445_at |
AT4G16750
|
[Integrase-type DNA-binding superfamily protein] |
-0.123 |
-0.078 |
| 153 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-0.119 |
-0.089 |
| 154 |
262412_at |
AT1G34760
|
GF14 OMICRON, GRF11, general regulatory factor 11, RHS5, ROOT HAIR SPECIFIC 5 |
-0.118 |
-0.123 |
| 155 |
264454_at |
AT1G10320
|
[Zinc finger C-x8-C-x5-C-x3-H type family protein] |
-0.117 |
0.031 |
| 156 |
259760_at |
AT1G77580
|
unknown |
-0.116 |
-0.174 |
| 157 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
-0.115 |
-0.318 |
| 158 |
264471_at |
AT1G67120
|
AtMDN1, MDN1, MIDASIN 1 |
-0.114 |
0.272 |
| 159 |
259448_at |
AT1G13790
|
FDM4, factor of DNA methylation 4 |
-0.112 |
0.120 |
| 160 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
-0.111 |
-0.214 |
| 161 |
264822_at |
AT1G03457
|
AtBRN2, BRN2, Bruno-like 2 |
-0.111 |
-0.102 |
| 162 |
252215_at |
AT3G50240
|
KICP-02 |
-0.108 |
-0.187 |
| 163 |
262586_at |
AT1G15480
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.107 |
0.143 |
| 164 |
267386_at |
AT2G44430
|
[DNA-binding bromodomain-containing protein] |
-0.106 |
-0.055 |
| 165 |
260664_at |
AT1G19510
|
ATRL5, RAD-like 5, RL5, RAD-like 5, RSM4, RADIALIS-LIKE SANT/MYB 4 |
-0.105 |
-0.042 |
| 166 |
249354_at |
AT5G40480
|
EMB3012, embryo defective 3012 |
-0.104 |
0.057 |
| 167 |
247005_at |
AT5G67520
|
adenosine-5'-phosphosulfate (APS) kinase 4 |
-0.103 |
0.183 |
| 168 |
250064_at |
AT5G17890
|
CHS3, CHILLING SENSITIVE 3, DAR4, DA1-related protein 4 |
-0.103 |
-0.079 |
| 169 |
250087_at |
AT5G17270
|
[Protein prenylyltransferase superfamily protein] |
-0.101 |
0.123 |
| 170 |
251402_at |
AT3G60290
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.101 |
-0.473 |
| 171 |
262336_at |
AT1G64220
|
TOM7-2, translocase of outer membrane 7 kDa subunit 2 |
-0.101 |
0.193 |
| 172 |
251023_at |
AT5G02170
|
[Transmembrane amino acid transporter family protein] |
-0.101 |
-0.020 |
| 173 |
253975_at |
AT4G26600
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.100 |
0.190 |
| 174 |
247305_at |
AT5G63905
|
unknown |
-0.100 |
0.040 |
| 175 |
263495_at |
AT2G42530
|
COR15B, cold regulated 15b |
-0.099 |
-0.261 |
| 176 |
246371_at |
AT1G51940
|
LYK3, LysM-containing receptor-like kinase 3 |
-0.098 |
-0.091 |
| 177 |
253034_at |
AT4G38230
|
ATCPK26, ARABIDOPSIS THALIANA CALCIUM-DEPENDENT PROTEIN KINASE 26, CPK26, calcium-dependent protein kinase 26 |
-0.096 |
-0.175 |
| 178 |
252520_at |
AT3G46370
|
[Leucine-rich repeat protein kinase family protein] |
-0.095 |
-0.063 |
| 179 |
247362_at |
AT5G63140
|
ATPAP29, purple acid phosphatase 29, PAP29, PAP29, purple acid phosphatase 29 |
-0.095 |
-0.067 |
| 180 |
253575_at |
AT4G31060
|
[Integrase-type DNA-binding superfamily protein] |
-0.095 |
-0.219 |
| 181 |
267519_at |
AT2G30470
|
HSI2, high-level expression of sugar-inducible gene 2, VAL1, VP1/ABI3-LIKE 1 |
-0.094 |
-0.051 |
| 182 |
261611_at |
AT1G49730
|
[Protein kinase superfamily protein] |
-0.094 |
-0.199 |
| 183 |
256919_at |
AT3G18970
|
MEF20, mitochondrial editing factor 20 |
-0.093 |
0.075 |
| 184 |
265349_at |
AT2G22610
|
[Di-glucose binding protein with Kinesin motor domain] |
-0.093 |
-0.104 |
| 185 |
247035_at |
AT5G67110
|
ALC, ALCATRAZ |
-0.093 |
0.000 |
| 186 |
261375_at |
AT1G53160
|
FTM6, FLORAL TRANSITION AT THE MERISTEM6, SPL4, squamosa promoter binding protein-like 4 |
-0.092 |
-0.162 |
| 187 |
261004_at |
AT1G26450
|
[Carbohydrate-binding X8 domain superfamily protein] |
-0.092 |
-0.099 |
| 188 |
257164_at |
AT3G24320
|
ATMSH1, CHM, CHLOROPLAST MUTATOR, CHM1, MSH1, MUTL protein homolog 1 |
-0.091 |
-0.018 |
| 189 |
267509_at |
AT2G45660
|
AGL20, AGAMOUS-like 20, ATSOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1, SOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1 |
-0.091 |
-0.081 |
| 190 |
253623_at |
AT4G30570
|
[Glucose-1-phosphate adenylyltransferase family protein] |
-0.089 |
-0.071 |
| 191 |
249113_at |
AT5G43790
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.089 |
-0.022 |
| 192 |
248788_at |
AT5G47430
|
[DWNN domain, a CCHC-type zinc finger] |
-0.089 |
-0.017 |
| 193 |
255264_at |
AT4G05170
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.089 |
0.043 |
| 194 |
253285_at |
AT4G34250
|
KCS16, 3-ketoacyl-CoA synthase 16 |
-0.088 |
0.009 |
| 195 |
253942_at |
AT4G27010
|
EMB2788, EMBRYO DEFECTIVE 2788 |
-0.088 |
0.171 |
| 196 |
264390_at |
AT1G11950
|
[Transcription factor jumonji (jmjC) domain-containing protein] |
-0.088 |
0.073 |
| 197 |
246929_at |
AT5G25210
|
unknown |
-0.088 |
0.072 |
| 198 |
259899_at |
AT1G71210
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.088 |
0.046 |
| 199 |
253663_at |
AT4G30160
|
ATVLN4, VLN4, villin 4 |
-0.088 |
-0.024 |
| 200 |
246572_at |
AT5G15010
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.087 |
0.025 |
| 201 |
247339_at |
AT5G63690
|
[Nucleic acid-binding, OB-fold-like protein] |
-0.086 |
0.077 |
| 202 |
265311_at |
AT2G20250
|
unknown |
-0.085 |
-0.215 |
| 203 |
257013_at |
AT3G26922
|
[F-box/RNI-like superfamily protein] |
-0.085 |
-0.140 |
| 204 |
253331_at |
AT4G33490
|
[Eukaryotic aspartyl protease family protein] |
-0.084 |
-0.284 |
| 205 |
254011_at |
AT4G26370
|
[antitermination NusB domain-containing protein] |
-0.084 |
-0.005 |
| 206 |
259585_at |
AT1G28090
|
[Polynucleotide adenylyltransferase family protein] |
-0.084 |
0.029 |
| 207 |
265877_at |
AT2G42380
|
ATBZIP34, BZIP34 |
-0.084 |
-0.269 |
| 208 |
250649_at |
AT5G06690
|
WCRKC1, WCRKC thioredoxin 1 |
-0.083 |
-0.043 |
| 209 |
248976_at |
AT5G44930
|
ARAD2, ARABINAN DEFICIENT 2 |
-0.083 |
0.055 |
| 210 |
250728_at |
AT5G06440
|
unknown |
-0.082 |
-0.106 |
| 211 |
250196_at |
AT5G14580
|
[polyribonucleotide nucleotidyltransferase, putative] |
-0.082 |
0.177 |
| 212 |
251956_at |
AT3G53460
|
CP29, chloroplast RNA-binding protein 29 |
-0.082 |
0.026 |
| 213 |
258260_at |
AT3G26850
|
[histone-lysine N-methyltransferases] |
-0.081 |
-0.008 |
| 214 |
252355_at |
AT3G48250
|
BIR6, Buthionine sulfoximine-insensitive roots 6 |
-0.081 |
0.037 |
| 215 |
266910_at |
AT2G45920
|
[U-box domain-containing protein] |
-0.081 |
-0.028 |
| 216 |
252586_at |
AT3G45610
|
DOF6, DOF transcription factor 6 |
-0.081 |
-0.158 |
| 217 |
246461_at |
AT5G16930
|
[AAA-type ATPase family protein] |
-0.080 |
0.194 |
| 218 |
250774_at |
AT5G05450
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.080 |
0.007 |
| 219 |
254665_at |
AT4G18340
|
[Glycosyl hydrolase superfamily protein] |
-0.080 |
0.356 |
| 220 |
254716_at |
AT4G13560
|
UNE15, unfertilized embryo sac 15 |
-0.080 |
-0.072 |
| 221 |
249124_at |
AT5G43740
|
[Disease resistance protein (CC-NBS-LRR class) family] |
-0.079 |
-0.059 |
| 222 |
255278_at |
AT4G04940
|
[transducin family protein / WD-40 repeat family protein] |
-0.078 |
0.222 |
| 223 |
263920_at |
AT2G36410
|
unknown |
-0.078 |
-0.022 |
| 224 |
249800_at |
AT5G23660
|
AtSWEET12, MTN3, homolog of Medicago truncatula MTN3, SWEET12 |
-0.078 |
0.087 |
| 225 |
257323_at |
ATMG01200
|
ORF294 |
-0.078 |
0.242 |
| 226 |
252649_at |
AT3G44680
|
AtHDA9, HDA09, HDA9, histone deacetylase 9 |
-0.078 |
0.021 |
| 227 |
251771_at |
AT3G56000
|
ATCSLA14, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE A14, CSLA14, cellulose synthase like A14 |
-0.077 |
-0.054 |
| 228 |
264806_at |
AT1G08610
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.077 |
0.160 |
| 229 |
250987_at |
AT5G02860
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.077 |
-0.054 |
| 230 |
251955_at |
AT3G53680
|
[Acyl-CoA N-acyltransferase with RING/FYVE/PHD-type zinc finger domain] |
-0.076 |
-0.117 |
| 231 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
-0.076 |
-0.076 |
| 232 |
258009_at |
AT3G19440
|
[Pseudouridine synthase family protein] |
-0.075 |
0.134 |
| 233 |
249059_at |
AT5G44530
|
[Subtilase family protein] |
-0.075 |
-0.317 |
| 234 |
258424_at |
AT3G16750
|
unknown |
-0.075 |
-0.056 |
| 235 |
254208_at |
AT4G24175
|
unknown |
-0.075 |
-0.049 |
| 236 |
249258_at |
AT5G41650
|
[Lactoylglutathione lyase / glyoxalase I family protein] |
-0.074 |
0.058 |
| 237 |
251256_at |
AT3G62300
|
unknown |
-0.074 |
-0.095 |
| 238 |
257554_at |
AT3G24890
|
ATVAMP728, VAMP728, vesicle-associated membrane protein 728 |
-0.074 |
0.010 |
| 239 |
248176_at |
AT5G54650
|
ATFH5, FORMIN HOMOLOGY 5, Fh5, formin homology5 |
-0.073 |
-0.029 |
| 240 |
261613_at |
AT1G49720
|
ABF1, abscisic acid responsive element-binding factor 1, AtABF1 |
-0.072 |
-0.048 |
| 241 |
263829_at |
AT2G40435
|
unknown |
-0.072 |
-0.472 |
| 242 |
267465_at |
AT2G19170
|
SLP3, subtilisin-like serine protease 3 |
-0.072 |
-0.287 |
| 243 |
259172_at |
AT3G03630
|
CS26, cysteine synthase 26 |
-0.071 |
0.013 |
| 244 |
245657_at |
AT1G56720
|
[Protein kinase superfamily protein] |
-0.071 |
-0.318 |
| 245 |
245029_at |
AT2G26580
|
YAB5, YABBY5 |
-0.070 |
-0.138 |
| 246 |
255070_at |
AT4G09020
|
ATISA3, ISA3, isoamylase 3 |
-0.070 |
-0.109 |
| 247 |
265854_at |
AT2G42370
|
unknown |
-0.070 |
-0.033 |
| 248 |
262512_at |
AT1G17145
|
[RING/U-box superfamily protein] |
-0.069 |
-0.025 |
| 249 |
260622_at |
AT1G07980
|
NF-YC10, nuclear factor Y, subunit C10 |
-0.069 |
0.022 |
| 250 |
264352_at |
AT1G03270
|
unknown |
-0.069 |
-0.121 |
| 251 |
264500_at |
AT1G09370
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.069 |
0.059 |
| 252 |
259287_at |
AT3G11490
|
[rac GTPase activating protein] |
-0.069 |
-0.052 |
| 253 |
256469_at |
AT1G32540
|
LOL1, lsd one like 1 |
-0.068 |
-0.038 |
| 254 |
245572_at |
AT4G14720
|
PPD2, PEAPOD 2, TIFY4B |
-0.068 |
-0.051 |
| 255 |
250529_at |
AT5G08610
|
PDE340, PIGMENT DEFECTIVE 340 |
-0.068 |
0.168 |
| 256 |
267349_at |
AT2G40010
|
[Ribosomal protein L10 family protein] |
-0.068 |
0.025 |
| 257 |
246999_at |
AT5G67440
|
MEL2, MAB4/ENP/NPY1-LIKE 2, NPY3, NAKED PINS IN YUC MUTANTS 3 |
-0.068 |
0.017 |
| 258 |
245405_at |
AT4G17150
|
[alpha/beta-Hydrolases superfamily protein] |
-0.068 |
-0.074 |
| 259 |
249953_at |
AT5G18960
|
FRS12, FAR1-related sequence 12 |
-0.067 |
-0.034 |
| 260 |
248201_at |
AT5G54180
|
PTAC15, plastid transcriptionally active 15 |
-0.067 |
0.026 |
| 261 |
252362_at |
AT3G48500
|
PDE312, PIGMENT DEFECTIVE 312, PTAC10, PLASTID TRANSCRIPTIONALLY ACTIVE 10, TAC10 |
-0.067 |
0.052 |
| 262 |
255617_at |
AT4G01330
|
[Protein kinase superfamily protein] |
-0.067 |
-0.118 |
| 263 |
254293_at |
AT4G23060
|
IQD22, IQ-domain 22 |
-0.066 |
-0.115 |
| 264 |
256784_at |
AT3G13674
|
unknown |
-0.066 |
-0.127 |
| 265 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
-0.066 |
-0.346 |
| 266 |
246231_at |
AT4G37080
|
unknown |
-0.066 |
-0.177 |
| 267 |
265154_at |
AT1G30960
|
[GTP-binding family protein] |
-0.065 |
0.305 |
| 268 |
249115_at |
AT5G43810
|
AGO10, ARGONAUTE 10, PNH, PINHEAD, ZLL, ZWILLE |
-0.065 |
-0.158 |
| 269 |
251225_at |
AT3G62660
|
GATL7, galacturonosyltransferase-like 7 |
-0.065 |
-0.080 |
| 270 |
266808_at |
AT2G29995
|
unknown |
-0.064 |
-0.049 |
| 271 |
254442_at |
AT4G21060
|
AtGALT2, GALT2, AGP galactosyltransferase 2 |
-0.064 |
-0.264 |
| 272 |
249551_at |
AT5G38220
|
[alpha/beta-Hydrolases superfamily protein] |
-0.063 |
-0.064 |
| 273 |
251966_at |
AT3G53380
|
LecRK-VIII.1, L-type lectin receptor kinase VIII.1 |
-0.063 |
-0.148 |
| 274 |
248705_at |
AT5G48520
|
AtAUG3, AUG3, augmin 3 |
-0.063 |
-0.128 |
| 275 |
249948_at |
AT5G19210
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.062 |
-0.077 |