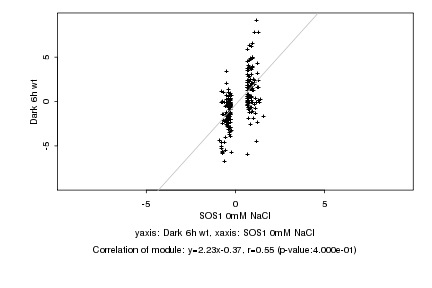
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
SOS1 0mM NaCl |
Log2 signal ratio
Dark 6h wt |
| 1 |
256527_at |
AT1G66100
|
[Plant thionin] |
1.560 |
-1.609 |
| 2 |
252462_at |
AT3G47250
|
unknown |
1.371 |
0.335 |
| 3 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
1.346 |
-0.047 |
| 4 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
1.269 |
2.434 |
| 5 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
1.258 |
0.193 |
| 6 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
1.258 |
7.790 |
| 7 |
245448_at |
AT4G16860
|
RPP4, recognition of peronospora parasitica 4 |
1.255 |
1.689 |
| 8 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
1.223 |
3.264 |
| 9 |
258225_at |
AT3G15630
|
unknown |
1.212 |
4.332 |
| 10 |
256021_at |
AT1G58270
|
ZW9 |
1.211 |
1.611 |
| 11 |
256497_at |
AT1G31580
|
CXC750, ECS1 |
1.211 |
-2.267 |
| 12 |
264958_at |
AT1G76960
|
unknown |
1.167 |
-4.501 |
| 13 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
1.138 |
-0.035 |
| 14 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
1.129 |
9.240 |
| 15 |
260312_at |
AT1G63880
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
1.101 |
0.407 |
| 16 |
252648_at |
AT3G44630
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
1.083 |
-1.277 |
| 17 |
265698_at |
AT2G32160
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
1.077 |
-0.766 |
| 18 |
245729_at |
AT1G73490
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
1.069 |
-0.230 |
| 19 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
1.064 |
7.840 |
| 20 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
1.055 |
2.022 |
| 21 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
1.047 |
2.152 |
| 22 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
1.027 |
2.460 |
| 23 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
1.001 |
-1.882 |
| 24 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.992 |
2.548 |
| 25 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.989 |
1.246 |
| 26 |
247543_at |
AT5G61600
|
ERF104, ethylene response factor 104 |
0.964 |
2.523 |
| 27 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
0.956 |
4.905 |
| 28 |
263023_at |
AT1G23960
|
unknown |
0.943 |
1.548 |
| 29 |
265918_at |
AT2G15090
|
KCS8, 3-ketoacyl-CoA synthase 8 |
0.938 |
2.051 |
| 30 |
259009_at |
AT3G09260
|
BGLU23, LEB, LONG ER BODY, PSR3.1, PYK10 |
0.932 |
-0.047 |
| 31 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.930 |
3.909 |
| 32 |
256617_at |
AT3G22240
|
unknown |
0.929 |
1.345 |
| 33 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
0.925 |
6.641 |
| 34 |
245749_at |
AT1G51090
|
[Heavy metal transport/detoxification superfamily protein ] |
0.924 |
5.050 |
| 35 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.921 |
4.033 |
| 36 |
248502_at |
AT5G50450
|
[HCP-like superfamily protein with MYND-type zinc finger] |
0.904 |
4.986 |
| 37 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
0.903 |
-1.105 |
| 38 |
253073_at |
AT4G37410
|
CYP81F4, cytochrome P450, family 81, subfamily F, polypeptide 4 |
0.883 |
-0.047 |
| 39 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
0.881 |
0.521 |
| 40 |
250063_at |
AT5G17880
|
CSA1, constitutive shade-avoidance1 |
0.874 |
0.123 |
| 41 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
0.871 |
6.283 |
| 42 |
255484_at |
AT4G02540
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.870 |
0.617 |
| 43 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
0.855 |
-0.577 |
| 44 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
0.851 |
3.652 |
| 45 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
0.850 |
3.107 |
| 46 |
251494_at |
AT3G59350
|
[Protein kinase superfamily protein] |
0.847 |
1.710 |
| 47 |
246253_at |
AT4G37260
|
ATMYB73, MYB73, myb domain protein 73 |
0.846 |
2.046 |
| 48 |
253485_at |
AT4G31800
|
ATWRKY18, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 18, WRKY18, WRKY DNA-binding protein 18 |
0.845 |
-1.158 |
| 49 |
264774_at |
AT1G22890
|
unknown |
0.840 |
1.410 |
| 50 |
260881_at |
AT1G21550
|
[Calcium-binding EF-hand family protein] |
0.836 |
2.528 |
| 51 |
264045_at |
AT2G22450
|
AtRIBA2, RIBA2, homolog of ribA 2 |
0.836 |
4.779 |
| 52 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.822 |
4.931 |
| 53 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.820 |
-2.547 |
| 54 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.816 |
1.810 |
| 55 |
262010_at |
AT1G35612
|
[expressed protein] |
0.805 |
1.707 |
| 56 |
247013_at |
AT5G67480
|
ATBT4, BT4, BTB and TAZ domain protein 4 |
0.796 |
3.741 |
| 57 |
258792_at |
AT3G04640
|
[glycine-rich protein] |
0.792 |
0.507 |
| 58 |
253643_at |
AT4G29780
|
unknown |
0.790 |
0.597 |
| 59 |
252703_at |
AT3G43740
|
[Leucine-rich repeat (LRR) family protein] |
0.790 |
-0.623 |
| 60 |
262452_at |
AT1G11210
|
unknown |
0.788 |
6.362 |
| 61 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
0.788 |
-0.080 |
| 62 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
0.782 |
2.274 |
| 63 |
261658_at |
AT1G50040
|
unknown |
0.764 |
2.311 |
| 64 |
247867_at |
AT5G57630
|
CIPK21, CBL-interacting protein kinase 21, SnRK3.4, SNF1-RELATED PROTEIN KINASE 3.4 |
0.764 |
4.655 |
| 65 |
252485_at |
AT3G46530
|
RPP13, RECOGNITION OF PERONOSPORA PARASITICA 13 |
0.761 |
-1.186 |
| 66 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
0.752 |
-0.858 |
| 67 |
251507_at |
AT3G59080
|
[Eukaryotic aspartyl protease family protein] |
0.750 |
0.605 |
| 68 |
246018_at |
AT5G10695
|
unknown |
0.741 |
-0.039 |
| 69 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
0.739 |
-0.771 |
| 70 |
246831_at |
AT5G26340
|
ATSTP13, SUGAR TRANSPORT PROTEIN 13, MSS1, STP13, SUGAR TRANSPORT PROTEIN 13 |
0.739 |
2.869 |
| 71 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
0.734 |
0.501 |
| 72 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
0.733 |
3.935 |
| 73 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.730 |
-0.897 |
| 74 |
248392_at |
AT5G52050
|
[MATE efflux family protein] |
0.720 |
-1.888 |
| 75 |
262458_at |
AT1G11280
|
[S-locus lectin protein kinase family protein] |
0.717 |
-0.209 |
| 76 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.717 |
3.527 |
| 77 |
255733_at |
AT1G25400
|
unknown |
0.716 |
4.162 |
| 78 |
260411_at |
AT1G69890
|
unknown |
0.715 |
2.878 |
| 79 |
257076_at |
AT3G19680
|
unknown |
0.708 |
0.567 |
| 80 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
0.707 |
2.402 |
| 81 |
246200_at |
AT4G37240
|
unknown |
0.706 |
-0.596 |
| 82 |
249242_at |
AT5G42250
|
[Zinc-binding alcohol dehydrogenase family protein] |
0.705 |
0.545 |
| 83 |
262698_at |
AT1G75960
|
[AMP-dependent synthetase and ligase family protein] |
0.702 |
0.787 |
| 84 |
266316_at |
AT2G27080
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.696 |
0.368 |
| 85 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
0.691 |
-0.658 |
| 86 |
255931_at |
AT1G12710
|
AtPP2-A12, phloem protein 2-A12, PP2-A12, phloem protein 2-A12 |
0.690 |
1.861 |
| 87 |
266246_at |
AT2G27690
|
CYP94C1, cytochrome P450, family 94, subfamily C, polypeptide 1 |
0.686 |
0.602 |
| 88 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
0.685 |
1.472 |
| 89 |
257476_at |
AT1G80960
|
[F-box and Leucine Rich Repeat domains containing protein] |
0.677 |
0.864 |
| 90 |
252679_at |
AT3G44260
|
AtCAF1a, CCR4- associated factor 1a, CAF1a, CCR4- associated factor 1a |
0.674 |
-0.281 |
| 91 |
262211_at |
AT1G74930
|
ORA47 |
0.668 |
3.123 |
| 92 |
252214_at |
AT3G50260
|
ATERF#011, CEJ1, cooperatively regulated by ethylene and jasmonate 1, DEAR1, DREB AND EAR MOTIF PROTEIN 1 |
0.667 |
3.766 |
| 93 |
266259_at |
AT2G27830
|
unknown |
0.663 |
3.540 |
| 94 |
264166_at |
AT1G65370
|
[TRAF-like family protein] |
0.658 |
0.015 |
| 95 |
247937_at |
AT5G57110
|
ACA8, autoinhibited Ca2+ -ATPase, isoform 8, AT-ACA8, AUTOINHIBITED CA2+ -ATPASE, ISOFORM 8 |
0.657 |
1.823 |
| 96 |
257592_at |
AT3G24982
|
ATRLP40, receptor like protein 40, RLP40, receptor like protein 40 |
0.653 |
0.154 |
| 97 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
0.648 |
2.152 |
| 98 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
0.646 |
5.877 |
| 99 |
265117_at |
AT1G62500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.645 |
-0.047 |
| 100 |
252053_at |
AT3G52400
|
ATSYP122, SYP122, syntaxin of plants 122 |
0.641 |
0.361 |
| 101 |
250777_at |
AT5G05440
|
PYL5, PYRABACTIN RESISTANCE 1-LIKE 5, RCAR8, regulatory component of ABA receptor 8 |
0.637 |
-0.431 |
| 102 |
250755_at |
AT5G05750
|
[DNAJ heat shock N-terminal domain-containing protein] |
0.635 |
1.313 |
| 103 |
253915_at |
AT4G27280
|
[Calcium-binding EF-hand family protein] |
0.632 |
0.855 |
| 104 |
245731_at |
AT1G73500
|
ATMKK9, MKK9, MAP kinase kinase 9 |
0.632 |
0.601 |
| 105 |
247524_at |
AT5G61440
|
ACHT5, atypical CYS HIS rich thioredoxin 5 |
0.632 |
1.662 |
| 106 |
260037_at |
AT1G68840
|
AtRAV2, EDF2, ETHYLENE RESPONSE DNA BINDING FACTOR 2, RAP2.8, RELATED TO AP2 8, RAV2, related to ABI3/VP1 2, TEM2, TEMPRANILLO 2 |
0.632 |
0.575 |
| 107 |
267349_at |
AT2G40010
|
[Ribosomal protein L10 family protein] |
0.632 |
-0.108 |
| 108 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
0.630 |
-5.882 |
| 109 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
0.628 |
2.433 |
| 110 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
0.628 |
-0.404 |
| 111 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
0.626 |
2.255 |
| 112 |
259364_at |
AT1G13260
|
EDF4, ETHYLENE RESPONSE DNA BINDING FACTOR 4, RAV1, related to ABI3/VP1 1 |
0.626 |
1.663 |
| 113 |
258939_at |
AT3G10020
|
unknown |
0.626 |
4.558 |
| 114 |
263574_at |
AT2G16990
|
[Major facilitator superfamily protein] |
0.625 |
1.821 |
| 115 |
245252_at |
AT4G17500
|
ATERF-1, ethylene responsive element binding factor 1, ERF-1, ethylene responsive element binding factor 1 |
0.625 |
1.654 |
| 116 |
265530_at |
AT2G06050
|
AtOPR3, DDE1, DELAYED DEHISCENCE 1, OPR3, oxophytodienoate-reductase 3 |
0.623 |
0.396 |
| 117 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
-0.895 |
-4.360 |
| 118 |
250135_at |
AT5G15360
|
unknown |
-0.830 |
-0.047 |
| 119 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-0.825 |
-4.984 |
| 120 |
258370_at |
AT3G14395
|
unknown |
-0.819 |
-4.510 |
| 121 |
250561_at |
AT5G08030
|
AtGDPD6, GDPD6, glycerophosphodiester phosphodiesterase 6 |
-0.806 |
-5.265 |
| 122 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.787 |
1.225 |
| 123 |
253309_at |
AT4G33790
|
CER4, ECERIFERUM 4, FAR3, FATTY ACID REDUCTASE 3, G7 |
-0.764 |
-5.611 |
| 124 |
254931_at |
AT4G11460
|
CRK30, cysteine-rich RLK (RECEPTOR-like protein kinase) 30 |
-0.759 |
-5.822 |
| 125 |
247225_at |
AT5G65090
|
BST1, BRISTLED 1, DER4, DEFORMED ROOT HAIRS 4, MRH3 |
-0.743 |
-2.417 |
| 126 |
254566_at |
AT4G19240
|
unknown |
-0.738 |
-0.047 |
| 127 |
262421_at |
AT1G50290
|
unknown |
-0.737 |
0.065 |
| 128 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
-0.734 |
-1.355 |
| 129 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
-0.730 |
-5.648 |
| 130 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
-0.719 |
-2.128 |
| 131 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
-0.676 |
-1.440 |
| 132 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-0.674 |
1.028 |
| 133 |
261544_at |
AT1G63540
|
[hydroxyproline-rich glycoprotein family protein] |
-0.649 |
-0.047 |
| 134 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
-0.648 |
-6.708 |
| 135 |
248372_at |
AT5G51850
|
TRM24, TON1 Recruiting Motif 24 |
-0.635 |
-2.144 |
| 136 |
263918_at |
AT2G36590
|
ATPROT3, PROLINE TRANSPORTER 3, ProT3, proline transporter 3 |
-0.633 |
-4.552 |
| 137 |
248246_at |
AT5G53200
|
TRY, TRIPTYCHON |
-0.609 |
-2.236 |
| 138 |
257451_at |
AT1G05690
|
BT3, BTB and TAZ domain protein 3 |
-0.604 |
-1.381 |
| 139 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
-0.599 |
-2.076 |
| 140 |
247447_at |
AT5G62730
|
[Major facilitator superfamily protein] |
-0.587 |
-5.419 |
| 141 |
254145_at |
AT4G24700
|
unknown |
-0.579 |
-3.978 |
| 142 |
266912_at |
AT2G45900
|
TRM13, TON1 Recruiting Motif 13 |
-0.574 |
-0.482 |
| 143 |
245341_at |
AT4G16447
|
unknown |
-0.550 |
-1.226 |
| 144 |
258315_at |
AT3G16175
|
[Thioesterase superfamily protein] |
-0.539 |
-2.143 |
| 145 |
260471_at |
AT1G11070
|
unknown |
-0.533 |
0.723 |
| 146 |
258082_at |
AT3G25905
|
CLE27, CLAVATA3/ESR-RELATED 27 |
-0.529 |
-1.534 |
| 147 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.529 |
0.254 |
| 148 |
259042_at |
AT3G03450
|
RGL2, RGA-like 2 |
-0.527 |
-2.481 |
| 149 |
262212_at |
AT1G74890
|
ARR15, response regulator 15 |
-0.526 |
-0.070 |
| 150 |
264887_at |
AT1G23120
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.513 |
2.137 |
| 151 |
264513_at |
AT1G09420
|
G6PD4, glucose-6-phosphate dehydrogenase 4 |
-0.510 |
3.423 |
| 152 |
261150_at |
AT1G19640
|
JMT, jasmonic acid carboxyl methyltransferase |
-0.504 |
-2.016 |
| 153 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.503 |
-1.642 |
| 154 |
247406_at |
AT5G62920
|
ARR6, response regulator 6 |
-0.502 |
-0.411 |
| 155 |
253166_at |
AT4G35290
|
ATGLR3.2, ATGLUR2, GLR3.2, GLUTAMATE RECEPTOR 3.2, GLUR2, glutamate receptor 2 |
-0.473 |
-1.411 |
| 156 |
264083_at |
AT2G31230
|
ATERF15, ethylene-responsive element binding factor 15, ERF15, ethylene-responsive element binding factor 15 |
-0.469 |
-2.779 |
| 157 |
264130_at |
AT1G79160
|
unknown |
-0.468 |
-0.042 |
| 158 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
-0.455 |
-1.849 |
| 159 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.454 |
-2.681 |
| 160 |
264916_at |
AT1G60810
|
ACLA-2, ATP-citrate lyase A-2 |
-0.447 |
-0.100 |
| 161 |
256873_at |
AT3G26310
|
CYP71B35, cytochrome P450, family 71, subfamily B, polypeptide 35 |
-0.445 |
-1.394 |
| 162 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
-0.442 |
0.648 |
| 163 |
258663_at |
AT3G08670
|
unknown |
-0.442 |
1.407 |
| 164 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
-0.433 |
-2.924 |
| 165 |
262529_at |
AT1G17250
|
AtRLP3, receptor like protein 3, RFO2, RESISTANCE TO FUSARIUM OXYSPORUM 2, RLP3, receptor like protein 3 |
-0.432 |
-0.047 |
| 166 |
258038_at |
AT3G21260
|
GLTP3, GLYCOLIPID TRANSFER PROTEIN 3 |
-0.429 |
0.019 |
| 167 |
265999_at |
AT2G24100
|
ASG1, ALTERED SEED GERMINATION 1 |
-0.426 |
-0.275 |
| 168 |
262737_at |
AT1G28560
|
SRD2, SHOOT REDIFFERENTIATION DEFECTIVE 2 |
-0.426 |
-2.045 |
| 169 |
256976_at |
AT3G21020
|
[copia-like retrotransposon family, has a 5.4e-14 P-value blast match to gb|AAO73529.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
-0.424 |
1.034 |
| 170 |
262196_at |
AT1G77870
|
MUB5, membrane-anchored ubiquitin-fold protein 5 precursor |
-0.419 |
-2.398 |
| 171 |
249336_at |
AT5G41070
|
DRB5, dsRNA-binding protein 5 |
-0.416 |
-1.806 |
| 172 |
245090_at |
AT2G40900
|
UMAMIT11, Usually multiple acids move in and out Transporters 11 |
-0.411 |
-1.255 |
| 173 |
265483_at |
AT2G15790
|
CYP40, CYCLOPHILIN 40, SQN, SQUINT |
-0.409 |
0.413 |
| 174 |
251090_at |
AT5G01340
|
AtmSFC1, mSFC1, mitochondrial succinate-fumarate carrier 1 |
-0.407 |
-0.033 |
| 175 |
266830_at |
AT2G22810
|
ACC4, 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE POLYPEPTIDE, ACS4, 1-aminocyclopropane-1-carboxylate synthase 4, ATACS4 |
-0.406 |
-0.589 |
| 176 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.405 |
-2.337 |
| 177 |
258677_at |
AT3G08730
|
ATPK1, ARABIDOPSIS THALIANA PROTEIN-SERINE KINASE 1, ATPK6, ARABIDOPSIS THALIANA PROTEIN-SERINE KINASE 6, ATS6K1, PK1, protein-serine kinase 1, PK6, ROTEIN-SERINE KINASE 6, S6K1, P70 RIBOSOMAL S6 KINASE |
-0.403 |
-0.542 |
| 178 |
260805_at |
AT1G78320
|
ATGSTU23, glutathione S-transferase TAU 23, GSTU23, glutathione S-transferase TAU 23 |
-0.402 |
-3.136 |
| 179 |
252594_at |
AT3G45680
|
[Major facilitator superfamily protein] |
-0.402 |
-1.220 |
| 180 |
251454_at |
AT3G60080
|
[RING/U-box superfamily protein] |
-0.379 |
-1.593 |
| 181 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.376 |
-3.452 |
| 182 |
247399_at |
AT5G62960
|
unknown |
-0.373 |
-0.617 |
| 183 |
255360_at |
AT4G03960
|
AtPFA-DSP4, PFA-DSP4, plant and fungi atypical dual-specificity phosphatase 4 |
-0.366 |
-0.005 |
| 184 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.365 |
-0.549 |
| 185 |
247255_at |
AT5G64780
|
[Uncharacterised conserved protein UCP009193] |
-0.363 |
0.766 |
| 186 |
248607_at |
AT5G49480
|
ATCP1, Ca2+-binding protein 1, CP1, Ca2+-binding protein 1 |
-0.356 |
-2.863 |
| 187 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.355 |
-1.600 |
| 188 |
264605_at |
AT1G04550
|
BDL, BODENLOS, IAA12, indole-3-acetic acid inducible 12 |
-0.353 |
-1.454 |
| 189 |
247264_at |
AT5G64530
|
ANAC104, Arabidopsis NAC domain containing protein 104, XND1, xylem NAC domain 1 |
-0.353 |
-0.530 |
| 190 |
251586_at |
AT3G58070
|
GIS, GLABROUS INFLORESCENCE STEMS |
-0.352 |
-3.678 |
| 191 |
261945_at |
AT1G64530
|
NLP6, NIN-like protein 6 |
-0.340 |
-0.877 |
| 192 |
258409_at |
AT3G17640
|
[Leucine-rich repeat (LRR) family protein] |
-0.334 |
-1.191 |
| 193 |
257959_at |
AT3G25560
|
AtNIK2, NIK2, NSP-interacting kinase 2 |
-0.332 |
-0.443 |
| 194 |
246193_at |
AT5G20880
|
[retrotransposon family] |
-0.329 |
-0.205 |
| 195 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
-0.312 |
-1.323 |
| 196 |
259761_at |
AT1G77590
|
LACS9, long chain acyl-CoA synthetase 9 |
-0.310 |
-1.374 |
| 197 |
260059_at |
AT1G78090
|
ATTPPB, Arabidopsis thaliana trehalose-6-phosphate phosphatase B, TPPB, trehalose-6-phosphate phosphatase B |
-0.309 |
0.990 |
| 198 |
260420_at |
AT1G69610
|
[FUNCTIONS IN: structural constituent of ribosome] |
-0.309 |
-1.959 |
| 199 |
267159_at |
AT2G37650
|
[GRAS family transcription factor] |
-0.307 |
-0.072 |
| 200 |
259359_at |
AT1G13460
|
[Protein phosphatase 2A regulatory B subunit family protein] |
-0.304 |
-0.300 |
| 201 |
267545_at |
AT2G32690
|
ATGRP23, GLYCINE-RICH PROTEIN 23, GRP23, glycine-rich protein 23 |
-0.302 |
-1.142 |
| 202 |
252167_at |
AT3G50560
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.302 |
-0.265 |
| 203 |
248484_at |
AT5G51030
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.302 |
-1.923 |
| 204 |
260840_at |
AT1G29050
|
TBL38, TRICHOME BIREFRINGENCE-LIKE 38 |
-0.300 |
0.419 |
| 205 |
247522_at |
AT5G61340
|
unknown |
-0.298 |
-0.347 |
| 206 |
257801_at |
AT3G18750
|
ATWNK6, ARABIDOPSIS THALIANA WITH NO K 6, WNK6, with no lysine (K) kinase 6, ZIK5 |
-0.297 |
-2.901 |
| 207 |
262604_at |
AT1G15060
|
[Uncharacterised conserved protein UCP031088, alpha/beta hydrolase] |
-0.295 |
-0.271 |
| 208 |
248291_at |
AT5G53020
|
[Ribonuclease P protein subunit P38-related] |
-0.295 |
-0.450 |
| 209 |
263248_at |
AT2G31450
|
ATNTH1 |
-0.294 |
-0.193 |
| 210 |
255150_at |
AT4G08160
|
[glycosyl hydrolase family 10 protein / carbohydrate-binding domain-containing protein] |
-0.294 |
-0.885 |
| 211 |
253246_at |
AT4G34600
|
unknown |
-0.291 |
-0.524 |
| 212 |
248537_at |
AT5G50100
|
[Putative thiol-disulphide oxidoreductase DCC] |
-0.289 |
-2.001 |
| 213 |
267359_at |
AT2G40020
|
[Nucleolar histone methyltransferase-related protein] |
-0.289 |
-1.924 |
| 214 |
264549_at |
AT1G09440
|
[Protein kinase superfamily protein] |
-0.289 |
-1.385 |
| 215 |
249579_at |
AT5G37680
|
ARLA1A, ADP-ribosylation factor-like A1A, ATARLA1A, ADP-ribosylation factor-like A1A |
-0.289 |
-0.004 |
| 216 |
254226_at |
AT4G23690
|
AtDIR6, Arabidopsis thaliana dirigent protein 6, DIR6, dirigent protein 6 |
-0.288 |
-2.818 |
| 217 |
260073_at |
AT1G73660
|
SIS8, SUGAR INSENSITIVE 8 |
-0.287 |
0.232 |
| 218 |
263382_at |
AT2G40230
|
[HXXXD-type acyl-transferase family protein] |
-0.287 |
-0.561 |
| 219 |
265726_at |
AT2G32010
|
CVL1, CVP2 like 1 |
-0.286 |
-0.418 |
| 220 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.285 |
-3.855 |
| 221 |
263473_at |
AT2G31750
|
UGT74D1, UDP-glucosyl transferase 74D1 |
-0.284 |
0.165 |
| 222 |
246021_at |
AT5G21100
|
[Plant L-ascorbate oxidase] |
-0.284 |
-3.293 |
| 223 |
247263_at |
AT5G64470
|
TBL12, TRICHOME BIREFRINGENCE-LIKE 12 |
-0.283 |
-0.319 |
| 224 |
257951_at |
AT3G21700
|
ATSGP2, SGP2 |
-0.282 |
0.400 |
| 225 |
247015_at |
AT5G66960
|
[Prolyl oligopeptidase family protein] |
-0.282 |
-0.599 |
| 226 |
249290_at |
AT5G41060
|
[DHHC-type zinc finger family protein] |
-0.281 |
-0.345 |
| 227 |
247415_at |
AT5G63060
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.279 |
-2.320 |
| 228 |
264102_at |
AT1G79270
|
ECT8, evolutionarily conserved C-terminal region 8 |
-0.278 |
-0.623 |
| 229 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
-0.277 |
-3.154 |
| 230 |
255088_at |
AT4G09350
|
CRRJ, CHLORORESPIRATORY REDUCTION J, DJC75, DNA J protein C75, NdhT, NADH dehydrogenase-like complex T |
-0.276 |
-5.690 |
| 231 |
254979_at |
AT4G10550
|
[Subtilase family protein] |
-0.275 |
0.737 |