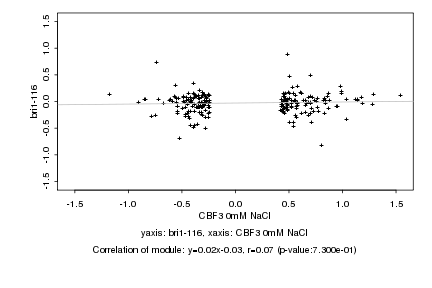
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
CBF3 0mM NaCl |
Log2 signal ratio
bri1-116 |
| 1 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
1.538 |
0.121 |
| 2 |
267459_at |
AT2G33850
|
unknown |
1.285 |
0.138 |
| 3 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
1.273 |
-0.054 |
| 4 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
1.183 |
-0.020 |
| 5 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
1.175 |
0.091 |
| 6 |
245749_at |
AT1G51090
|
[Heavy metal transport/detoxification superfamily protein ] |
1.136 |
0.028 |
| 7 |
263789_at |
AT2G24560
|
[GDSL-like Lipase/Acylhydrolase family protein] |
1.133 |
0.052 |
| 8 |
258225_at |
AT3G15630
|
unknown |
1.120 |
0.049 |
| 9 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
1.036 |
0.054 |
| 10 |
264511_at |
AT1G09350
|
AtGolS3, galactinol synthase 3, GolS3, galactinol synthase 3 |
1.031 |
-0.323 |
| 11 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
0.988 |
0.193 |
| 12 |
265918_at |
AT2G15090
|
KCS8, 3-ketoacyl-CoA synthase 8 |
0.987 |
0.163 |
| 13 |
260264_at |
AT1G68500
|
unknown |
0.972 |
0.294 |
| 14 |
246922_at |
AT5G25110
|
CIPK25, CBL-interacting protein kinase 25, SnRK3.25, SNF1-RELATED PROTEIN KINASE 3.25 |
0.947 |
-0.084 |
| 15 |
261658_at |
AT1G50040
|
unknown |
0.937 |
-0.092 |
| 16 |
251229_at |
AT3G62740
|
BGLU7, beta glucosidase 7 |
0.871 |
0.033 |
| 17 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
0.868 |
-0.126 |
| 18 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
0.862 |
0.167 |
| 19 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.855 |
0.109 |
| 20 |
247280_at |
AT5G64260
|
EXL2, EXORDIUM like 2 |
0.834 |
-0.037 |
| 21 |
263495_at |
AT2G42530
|
COR15B, cold regulated 15b |
0.833 |
0.031 |
| 22 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
0.830 |
0.057 |
| 23 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
0.826 |
-0.211 |
| 24 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
0.815 |
0.031 |
| 25 |
255608_at |
AT4G01140
|
unknown |
0.800 |
-0.815 |
| 26 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
0.768 |
-0.180 |
| 27 |
258893_at |
AT3G05660
|
AtRLP33, receptor like protein 33, RLP33, receptor like protein 33 |
0.763 |
0.072 |
| 28 |
263979_at |
AT2G42840
|
PDF1, protodermal factor 1 |
0.759 |
-0.101 |
| 29 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.756 |
0.010 |
| 30 |
266259_at |
AT2G27830
|
unknown |
0.730 |
0.025 |
| 31 |
248466_at |
AT5G50720
|
ATHVA22E, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE E, HVA22E, HVA22 homologue E |
0.727 |
-0.171 |
| 32 |
255064_at |
AT4G08950
|
EXO, EXORDIUM |
0.712 |
0.079 |
| 33 |
252997_at |
AT4G38400
|
ATEXLA2, expansin-like A2, ATEXPL2, ATHEXP BETA 2.2, EXLA2, expansin-like A2, EXPL2, EXPANSIN L2 |
0.709 |
-0.380 |
| 34 |
246911_at |
AT5G25810
|
tny, TINY |
0.707 |
-0.115 |
| 35 |
246253_at |
AT4G37260
|
ATMYB73, MYB73, myb domain protein 73 |
0.701 |
0.105 |
| 36 |
252006_at |
AT3G52820
|
ATPAP22, PURPLE ACID PHOSPHATASE 22, PAP22, purple acid phosphatase 22 |
0.699 |
-0.018 |
| 37 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
0.699 |
-0.029 |
| 38 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
0.693 |
-0.212 |
| 39 |
247013_at |
AT5G67480
|
ATBT4, BT4, BTB and TAZ domain protein 4 |
0.693 |
0.495 |
| 40 |
253485_at |
AT4G31800
|
ATWRKY18, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 18, WRKY18, WRKY DNA-binding protein 18 |
0.692 |
-0.035 |
| 41 |
265119_at |
AT1G62570
|
FMO GS-OX4, flavin-monooxygenase glucosinolate S-oxygenase 4 |
0.678 |
-0.248 |
| 42 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
0.678 |
0.084 |
| 43 |
264246_at |
AT1G60140
|
ATTPS10, trehalose phosphate synthase, TPS10, trehalose phosphate synthase, TPS10, TREHALOSE PHOSPHATE SYNTHASE 10 |
0.675 |
-0.009 |
| 44 |
259606_at |
AT1G27920
|
MAP65-8, microtubule-associated protein 65-8 |
0.660 |
-0.020 |
| 45 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
0.650 |
-0.198 |
| 46 |
257076_at |
AT3G19680
|
unknown |
0.650 |
0.027 |
| 47 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
0.647 |
-0.067 |
| 48 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
0.629 |
0.025 |
| 49 |
248870_at |
AT5G46710
|
[PLATZ transcription factor family protein] |
0.614 |
0.162 |
| 50 |
266140_at |
AT2G28120
|
[Major facilitator superfamily protein] |
0.613 |
0.156 |
| 51 |
259570_at |
AT1G20440
|
AtCOR47, COR47, cold-regulated 47, RD17 |
0.611 |
-0.223 |
| 52 |
251373_at |
AT3G60530
|
GATA4, GATA transcription factor 4 |
0.603 |
0.186 |
| 53 |
247776_at |
AT5G58700
|
ATPLC4, PLC4, phosphatidylinositol-speciwc phospholipase C4 |
0.577 |
-0.006 |
| 54 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
0.577 |
-0.031 |
| 55 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
0.577 |
0.300 |
| 56 |
264652_at |
AT1G08920
|
ESL1, ERD (early response to dehydration) six-like 1 |
0.575 |
-0.070 |
| 57 |
249601_at |
AT5G37980
|
[Zinc-binding dehydrogenase family protein] |
0.565 |
-0.080 |
| 58 |
264989_at |
AT1G27200
|
unknown |
0.565 |
-0.124 |
| 59 |
246200_at |
AT4G37240
|
unknown |
0.563 |
-0.289 |
| 60 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
0.558 |
-0.249 |
| 61 |
259364_at |
AT1G13260
|
EDF4, ETHYLENE RESPONSE DNA BINDING FACTOR 4, RAV1, related to ABI3/VP1 1 |
0.558 |
0.033 |
| 62 |
247540_at |
AT5G61590
|
[Integrase-type DNA-binding superfamily protein] |
0.557 |
-0.017 |
| 63 |
266545_at |
AT2G35290
|
SAUR79, SMALL AUXIN UPREGULATED RNA 79 |
0.556 |
0.122 |
| 64 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
0.551 |
0.018 |
| 65 |
265481_at |
AT2G15960
|
unknown |
0.546 |
0.026 |
| 66 |
251221_at |
AT3G62550
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.542 |
0.157 |
| 67 |
261926_at |
AT1G22530
|
PATL2, PATELLIN 2 |
0.539 |
-0.376 |
| 68 |
251768_at |
AT3G55940
|
[Phosphoinositide-specific phospholipase C family protein] |
0.537 |
-0.461 |
| 69 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
0.527 |
0.265 |
| 70 |
251356_at |
AT3G61060
|
AtPP2-A13, phloem protein 2-A13, PP2-A13, phloem protein 2-A13 |
0.523 |
-0.087 |
| 71 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
0.523 |
0.035 |
| 72 |
247189_at |
AT5G65390
|
AGP7, arabinogalactan protein 7 |
0.518 |
-0.103 |
| 73 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
0.515 |
-0.030 |
| 74 |
267209_at |
AT2G30930
|
unknown |
0.503 |
-0.389 |
| 75 |
256098_at |
AT1G13700
|
PGL1, 6-phosphogluconolactonase 1 |
0.501 |
0.166 |
| 76 |
253811_at |
AT4G28190
|
ULT, ULTRAPETALA, ULT1, ULTRAPETALA1 |
0.499 |
0.480 |
| 77 |
264482_at |
AT1G77210
|
AtSTP14, sugar transport protein 14, STP14, sugar transport protein 14 |
0.496 |
0.059 |
| 78 |
263421_at |
AT2G17230
|
EXL5, EXORDIUM like 5 |
0.492 |
0.177 |
| 79 |
252127_at |
AT3G50960
|
PLP3a, phosducin-like protein 3 homolog |
0.486 |
-0.040 |
| 80 |
266799_at |
AT2G22860
|
ATPSK2, phytosulfokine 2 precursor, PSK2, phytosulfokine 2 precursor |
0.484 |
-0.167 |
| 81 |
256633_at |
AT3G28340
|
GATL10, galacturonosyltransferase-like 10, GolS8, galactinol synthase 8 |
0.484 |
-0.147 |
| 82 |
253285_at |
AT4G34250
|
KCS16, 3-ketoacyl-CoA synthase 16 |
0.482 |
0.050 |
| 83 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.479 |
0.890 |
| 84 |
258589_at |
AT3G04290
|
ATLTL1, LTL1, Li-tolerant lipase 1 |
0.478 |
-0.058 |
| 85 |
247543_at |
AT5G61600
|
ERF104, ethylene response factor 104 |
0.475 |
0.019 |
| 86 |
252956_at |
AT4G38580
|
ATFP6, farnesylated protein 6, FP6, farnesylated protein 6, HIPP26, HEAVY METAL ASSOCIATED ISOPRENYLATED PLANT PROTEIN 26 |
0.474 |
0.023 |
| 87 |
246926_at |
AT5G25240
|
unknown |
0.474 |
-0.097 |
| 88 |
265170_at |
AT1G23730
|
ATBCA3, BETA CARBONIC ANHYDRASE 3, BCA3, beta carbonic anhydrase 3 |
0.472 |
-0.112 |
| 89 |
246756_at |
AT5G27930
|
[Protein phosphatase 2C family protein] |
0.468 |
-0.021 |
| 90 |
251753_at |
AT3G55760
|
unknown |
0.467 |
0.060 |
| 91 |
255381_at |
AT4G03510
|
ATRMA1, RMA1, RING membrane-anchor 1 |
0.465 |
0.024 |
| 92 |
252573_at |
AT3G45260
|
[C2H2-like zinc finger protein] |
0.464 |
0.156 |
| 93 |
251459_at |
AT3G60200
|
unknown |
0.456 |
0.137 |
| 94 |
249626_at |
AT5G37540
|
[Eukaryotic aspartyl protease family protein] |
0.456 |
0.110 |
| 95 |
250279_at |
AT5G13200
|
[GRAM domain family protein] |
0.455 |
-0.217 |
| 96 |
267393_at |
AT2G44500
|
[O-fucosyltransferase family protein] |
0.454 |
-0.118 |
| 97 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
0.452 |
0.033 |
| 98 |
246195_at |
AT4G36410
|
UBC17, ubiquitin-conjugating enzyme 17 |
0.448 |
-0.079 |
| 99 |
264521_at |
AT1G10020
|
unknown |
0.447 |
0.002 |
| 100 |
251192_at |
AT3G62720
|
ATXT1, XT1, xylosyltransferase 1, XXT1, XYG XYLOSYLTRANSFERASE 1 |
0.445 |
0.100 |
| 101 |
245794_at |
AT1G32170
|
XTH30, xyloglucan endotransglucosylase/hydrolase 30, XTR4, xyloglucan endotransglycosylase 4 |
0.444 |
-0.192 |
| 102 |
253880_at |
AT4G27590
|
[Heavy metal transport/detoxification superfamily protein ] |
0.444 |
0.004 |
| 103 |
258792_at |
AT3G04640
|
[glycine-rich protein] |
0.443 |
-0.173 |
| 104 |
255652_at |
AT4G00950
|
MEE47, maternal effect embryo arrest 47 |
0.441 |
0.153 |
| 105 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
0.437 |
-0.013 |
| 106 |
248100_at |
AT5G55180
|
[O-Glycosyl hydrolases family 17 protein] |
0.436 |
-0.181 |
| 107 |
257237_at |
AT3G14890
|
[phosphoesterase] |
0.435 |
-0.131 |
| 108 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
0.435 |
-0.039 |
| 109 |
263128_at |
AT1G78600
|
BBX22, B-box domain protein 22, DBB3, DOUBLE B-BOX 3, LZF1, light-regulated zinc finger protein 1, STH3, SALT TOLERANCE HOMOLOG 3 |
0.430 |
-0.017 |
| 110 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
0.427 |
-0.052 |
| 111 |
248419_at |
AT5G51550
|
EXL3, EXORDIUM like 3 |
0.425 |
-0.002 |
| 112 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
0.421 |
0.047 |
| 113 |
246114_at |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
0.421 |
-0.086 |
| 114 |
251927_at |
AT3G53990
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.419 |
-0.168 |
| 115 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
-1.180 |
0.137 |
| 116 |
258370_at |
AT3G14395
|
unknown |
-0.914 |
-0.009 |
| 117 |
245465_at |
AT4G16590
|
ATCSLA01, cellulose synthase-like A01, ATCSLA1, CELLULOSE SYNTHASE-LIKE A1, CSLA01, CSLA01, cellulose synthase-like A01 |
-0.851 |
0.040 |
| 118 |
256874_at |
AT3G26320
|
CYP71B36, cytochrome P450, family 71, subfamily B, polypeptide 36 |
-0.842 |
0.051 |
| 119 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
-0.787 |
-0.265 |
| 120 |
261247_at |
AT1G20070
|
unknown |
-0.751 |
-0.250 |
| 121 |
261684_at |
AT1G47400
|
unknown |
-0.745 |
0.741 |
| 122 |
250485_at |
AT5G09990
|
PROPEP5, elicitor peptide 5 precursor |
-0.727 |
0.054 |
| 123 |
247447_at |
AT5G62730
|
[Major facilitator superfamily protein] |
-0.677 |
-0.020 |
| 124 |
253940_at |
AT4G26950
|
unknown |
-0.622 |
0.027 |
| 125 |
246429_at |
AT5G17450
|
HIPP21, heavy metal associated isoprenylated plant protein 21 |
-0.612 |
0.042 |
| 126 |
247055_at |
AT5G66740
|
unknown |
-0.594 |
0.013 |
| 127 |
257365_x_at |
AT2G26020
|
PDF1.2b, plant defensin 1.2b |
-0.572 |
0.111 |
| 128 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
-0.567 |
0.085 |
| 129 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
-0.562 |
0.311 |
| 130 |
264083_at |
AT2G31230
|
ATERF15, ethylene-responsive element binding factor 15, ERF15, ethylene-responsive element binding factor 15 |
-0.554 |
0.066 |
| 131 |
250408_at |
AT5G10930
|
CIPK5, CBL-interacting protein kinase 5, SnRK3.24, SNF1-RELATED PROTEIN KINASE 3.24 |
-0.551 |
-0.012 |
| 132 |
245736_at |
AT1G73330
|
ATDR4, drought-repressed 4, DR4, drought-repressed 4 |
-0.546 |
-0.170 |
| 133 |
262811_at |
AT1G11700
|
unknown |
-0.546 |
-0.091 |
| 134 |
254693_at |
AT4G17880
|
MYC4 |
-0.546 |
-0.075 |
| 135 |
263486_at |
AT2G22200
|
[Integrase-type DNA-binding superfamily protein] |
-0.544 |
-0.220 |
| 136 |
258807_at |
AT3G04030
|
MYR2 |
-0.540 |
0.062 |
| 137 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-0.529 |
-0.680 |
| 138 |
260693_at |
AT1G32450
|
AtNPF7.3, NPF7.3, NRT1/ PTR family 7.3, NRT1.5, nitrate transporter 1.5 |
-0.495 |
-0.116 |
| 139 |
254301_at |
AT4G22790
|
[MATE efflux family protein] |
-0.494 |
0.111 |
| 140 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
-0.490 |
0.082 |
| 141 |
247684_at |
AT5G59670
|
[Leucine-rich repeat protein kinase family protein] |
-0.487 |
0.010 |
| 142 |
259042_at |
AT3G03450
|
RGL2, RGA-like 2 |
-0.476 |
-0.006 |
| 143 |
255575_at |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
-0.475 |
-0.111 |
| 144 |
258270_at |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
-0.472 |
-0.273 |
| 145 |
255028_at |
AT4G09890
|
unknown |
-0.471 |
-0.241 |
| 146 |
264016_at |
AT2G21220
|
SAUR12, SMALL AUXIN UPREGULATED RNA 12 |
-0.461 |
0.080 |
| 147 |
260491_at |
AT1G51440
|
DALL2, DAD1-Like Lipase 2 |
-0.459 |
0.020 |
| 148 |
249136_at |
AT5G43180
|
unknown |
-0.457 |
-0.044 |
| 149 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
-0.449 |
-0.222 |
| 150 |
246620_at |
AT5G36220
|
CYP81D1, cytochrome P450, family 81, subfamily D, polypeptide 1, CYP91A1, CYTOCHROME P450 91A1 |
-0.444 |
-0.184 |
| 151 |
265161_at |
AT1G30900
|
BP80-3;3, binding protein of 80 kDa 3;3, VSR3;3, VACUOLAR SORTING RECEPTOR 3;3, VSR6, VACUOLAR SORTING RECEPTOR 6 |
-0.443 |
-0.263 |
| 152 |
249047_at |
AT5G44410
|
[FAD-binding Berberine family protein] |
-0.439 |
0.053 |
| 153 |
260419_at |
AT1G69730
|
[Wall-associated kinase family protein] |
-0.439 |
0.153 |
| 154 |
256529_at |
AT1G33260
|
[Protein kinase superfamily protein] |
-0.434 |
-0.308 |
| 155 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
-0.428 |
-0.444 |
| 156 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
-0.425 |
-0.174 |
| 157 |
251301_at |
AT3G61880
|
CYP78A9, cytochrome p450 78a9 |
-0.425 |
0.012 |
| 158 |
247602_at |
AT5G60900
|
RLK1, receptor-like protein kinase 1 |
-0.421 |
-0.006 |
| 159 |
250744_at |
AT5G05840
|
unknown |
-0.421 |
0.012 |
| 160 |
265828_at |
AT2G14520
|
unknown |
-0.421 |
0.076 |
| 161 |
266357_at |
AT2G32290
|
BAM6, beta-amylase 6, BMY5, BETA-AMYLASE 5 |
-0.416 |
-0.006 |
| 162 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
-0.416 |
0.035 |
| 163 |
261758_at |
AT1G08250
|
ADT6, arogenate dehydratase 6, AtADT6, Arabidopsis thaliana arogenate dehydratase 6 |
-0.408 |
-0.088 |
| 164 |
262935_at |
AT1G79410
|
AtOCT5, organic cation/carnitine transporter5, OCT5, organic cation/carnitine transporter5 |
-0.401 |
0.063 |
| 165 |
254377_at |
AT4G21650
|
[Subtilase family protein] |
-0.398 |
0.161 |
| 166 |
247252_at |
AT5G64770
|
CLEL 9, CLE-like 9, GLV2, GOLVEN 2, RGF9, root meristem growth factor 9 |
-0.394 |
-0.479 |
| 167 |
247549_at |
AT5G61420
|
AtMYB28, HAG1, HIGH ALIPHATIC GLUCOSINOLATE 1, MYB28, myb domain protein 28, PMG1, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 1 |
-0.392 |
0.339 |
| 168 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
-0.391 |
-0.181 |
| 169 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-0.390 |
0.148 |
| 170 |
266962_at |
AT2G39435
|
TRM18, TON1 Recruiting Motif 18 |
-0.390 |
-0.431 |
| 171 |
262396_at |
AT1G49470
|
unknown |
-0.385 |
0.084 |
| 172 |
256100_at |
AT1G13750
|
[Purple acid phosphatases superfamily protein] |
-0.383 |
-0.038 |
| 173 |
260420_at |
AT1G69610
|
[FUNCTIONS IN: structural constituent of ribosome] |
-0.381 |
0.119 |
| 174 |
259432_at |
AT1G01520
|
ASG4, ALTERED SEED GERMINATION 4 |
-0.372 |
0.095 |
| 175 |
257748_at |
AT3G18710
|
ATPUB29, ARABIDOPSIS THALIANA PLANT U-BOX 29, PUB29, plant U-box 29 |
-0.369 |
-0.106 |
| 176 |
251594_at |
AT3G57630
|
[exostosin family protein] |
-0.365 |
-0.105 |
| 177 |
262590_at |
AT1G15100
|
RHA2A, RING-H2 finger A2A |
-0.363 |
-0.106 |
| 178 |
259736_at |
AT1G64390
|
AtGH9C2, glycosyl hydrolase 9C2, GH9C2, glycosyl hydrolase 9C2 |
-0.355 |
-0.422 |
| 179 |
251005_at |
AT5G02590
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.354 |
0.122 |
| 180 |
256569_at |
AT3G19550
|
unknown |
-0.353 |
0.058 |
| 181 |
253246_at |
AT4G34600
|
unknown |
-0.353 |
-0.028 |
| 182 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
-0.350 |
-0.104 |
| 183 |
259743_at |
AT1G71140
|
[MATE efflux family protein] |
-0.349 |
-0.101 |
| 184 |
254119_at |
AT4G24780
|
[Pectin lyase-like superfamily protein] |
-0.342 |
-0.105 |
| 185 |
256299_at |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
-0.339 |
-0.189 |
| 186 |
263112_at |
AT1G03080
|
NET1D, Networked 1D |
-0.339 |
-0.003 |
| 187 |
255503_at |
AT4G02420
|
LecRK-IV.4, L-type lectin receptor kinase IV.4 |
-0.338 |
0.061 |
| 188 |
246989_at |
AT5G67350
|
unknown |
-0.338 |
0.216 |
| 189 |
266372_at |
AT2G41310
|
ARR8, RESPONSE REGULATOR 8, ATRR3, response regulator 3, RR3, response regulator 3 |
-0.330 |
-0.056 |
| 190 |
254320_at |
AT4G22580
|
[Exostosin family protein] |
-0.325 |
-0.096 |
| 191 |
264998_at |
AT1G67330
|
unknown |
-0.321 |
-0.229 |
| 192 |
246260_at |
AT1G31820
|
PUT1, POLYAMINE UPTAKE TRANSPORTER 1 |
-0.321 |
0.076 |
| 193 |
247604_at |
AT5G60950
|
COBL5, COBRA-like protein 5 precursor |
-0.319 |
-0.000 |
| 194 |
264901_at |
AT1G23090
|
AST91, sulfate transporter 91, SULTR3;3 |
-0.319 |
0.083 |
| 195 |
249754_at |
AT5G24530
|
DMR6, DOWNY MILDEW RESISTANT 6 |
-0.318 |
-0.188 |
| 196 |
253580_at |
AT4G30400
|
[RING/U-box superfamily protein] |
-0.316 |
-0.255 |
| 197 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
-0.315 |
-0.024 |
| 198 |
251131_at |
AT5G01190
|
LAC10, laccase 10 |
-0.314 |
-0.105 |
| 199 |
247980_at |
AT5G56860
|
GATA21, GATA TRANSCRIPTION FACTOR 21, GNC, GATA, nitrate-inducible, carbon metabolism-involved |
-0.310 |
0.181 |
| 200 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
-0.308 |
0.128 |
| 201 |
245571_at |
AT4G14695
|
[Uncharacterised protein family (UPF0041)] |
-0.306 |
-0.063 |
| 202 |
262507_at |
AT1G11330
|
[S-locus lectin protein kinase family protein] |
-0.299 |
0.146 |
| 203 |
246168_at |
AT5G32460
|
[Transcriptional factor B3 family protein] |
-0.295 |
0.149 |
| 204 |
253836_at |
AT4G27840
|
[SNARE-like superfamily protein] |
-0.293 |
0.020 |
| 205 |
245599_at |
AT4G14220
|
RHF1A, RING-H2 group F1A |
-0.292 |
-0.005 |
| 206 |
254416_at |
AT4G21380
|
ARK3, receptor kinase 3, RK3, receptor kinase 3 |
-0.290 |
-0.057 |
| 207 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
-0.288 |
0.112 |
| 208 |
246235_at |
AT4G36830
|
HOS3-1 |
-0.284 |
-0.168 |
| 209 |
262646_at |
AT1G62800
|
ASP4, aspartate aminotransferase 4 |
-0.284 |
-0.290 |
| 210 |
263972_at |
AT2G42760
|
unknown |
-0.282 |
-0.033 |
| 211 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
-0.280 |
-0.497 |
| 212 |
262705_at |
AT1G16260
|
[Wall-associated kinase family protein] |
-0.278 |
0.086 |
| 213 |
248160_at |
AT5G54470
|
BBX29, B-box domain protein 29 |
-0.278 |
0.027 |
| 214 |
248484_at |
AT5G51030
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.273 |
0.012 |
| 215 |
263032_at |
AT1G23850
|
unknown |
-0.273 |
0.065 |
| 216 |
259535_at |
AT1G12280
|
SUMM2, suppressor of mkk1 mkk2 2 |
-0.260 |
0.148 |
| 217 |
264052_at |
AT2G22330
|
CYP79B3, cytochrome P450, family 79, subfamily B, polypeptide 3 |
-0.259 |
-0.027 |
| 218 |
267101_at |
AT2G41480
|
AtPRX25, PRX25, peroxidase 25 |
-0.257 |
-0.293 |
| 219 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
-0.255 |
-0.110 |
| 220 |
251586_at |
AT3G58070
|
GIS, GLABROUS INFLORESCENCE STEMS |
-0.255 |
0.119 |
| 221 |
255125_at |
AT4G08250
|
[GRAS family transcription factor] |
-0.253 |
-0.057 |
| 222 |
262137_at |
AT1G77920
|
TGA7, TGACG sequence-specific binding protein 7 |
-0.252 |
-0.213 |
| 223 |
263796_at |
AT2G24540
|
AFR, ATTENUATED FAR-RED RESPONSE |
-0.251 |
0.020 |
| 224 |
253534_at |
AT4G31500
|
ATR4, ALTERED TRYPTOPHAN REGULATION 4, CYP83B1, cytochrome P450, family 83, subfamily B, polypeptide 1, RED1, RED ELONGATED 1, RNT1, RUNT 1, SUR2, SUPERROOT 2 |
-0.249 |
-0.197 |
| 225 |
255845_at |
AT2G33600
|
AtCRL2, CRL2, CCR(Cinnamoyl coA:NADP oxidoreductase)-like 2 |
-0.246 |
-0.018 |
| 226 |
260146_at |
AT1G52770
|
[Phototropic-responsive NPH3 family protein] |
-0.245 |
0.128 |
| 227 |
261335_at |
AT1G44800
|
SIAR1, Siliques Are Red 1, UMAMIT18, Usually multiple acids move in and out Transporters 18 |
-0.243 |
-0.111 |