|
probeID |
AGICode |
Annotation |
Log2 signal ratio
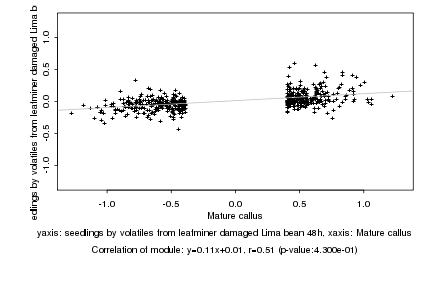
Mature callus |
Log2 signal ratio
seedlings by volatiles from leafminer damaged Lima bean 48h |
| 1 |
262325_at |
AT1G64160
|
AtDIR5, DIR5, dirigent protein 5 |
1.216 |
0.081 |
| 2 |
266141_at |
AT2G02120
|
LCR70, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 70, PDF2.1 |
1.059 |
0.038 |
| 3 |
266353_at |
AT2G01520
|
MLP328, MLP-like protein 328, ZCE1, (Zusammen-CA)-enhanced 1 |
1.056 |
-0.031 |
| 4 |
245267_at |
AT4G14060
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
1.031 |
-0.003 |
| 5 |
251531_at |
AT3G58550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.024 |
0.044 |
| 6 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
0.999 |
0.311 |
| 7 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
0.968 |
0.257 |
| 8 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
0.936 |
0.384 |
| 9 |
247359_at |
AT5G63560
|
FACT, FATTY ALCOHOL:CAFFEOYL-CoA CAFFEOYL TRANSFERASE |
0.924 |
0.035 |
| 10 |
248727_at |
AT5G47990
|
CYP705A5, cytochrome P450, family 705, subfamily A, polypeptide 5, THAD, THALIAN-DIOL DESATURASE, THAD1, THALIAN-DIOL DESATURASE |
0.919 |
0.006 |
| 11 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.914 |
0.124 |
| 12 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
0.914 |
0.171 |
| 13 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.908 |
0.417 |
| 14 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
0.905 |
0.208 |
| 15 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
0.885 |
0.176 |
| 16 |
247729_at |
AT5G59530
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.863 |
0.097 |
| 17 |
249599_at |
AT5G37990
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.853 |
0.042 |
| 18 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.850 |
0.092 |
| 19 |
258983_at |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
0.834 |
0.461 |
| 20 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
0.830 |
0.408 |
| 21 |
250302_at |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
0.827 |
-0.004 |
| 22 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
0.821 |
0.267 |
| 23 |
245393_at |
AT4G16260
|
[Glycosyl hydrolase superfamily protein] |
0.808 |
0.226 |
| 24 |
262608_at |
AT1G14120
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.807 |
-0.075 |
| 25 |
247358_at |
AT5G63580
|
ATFLS2, FLS2, flavonol synthase 2 |
0.800 |
0.212 |
| 26 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.795 |
0.132 |
| 27 |
265334_at |
AT2G18370
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.781 |
0.035 |
| 28 |
267548_at |
AT2G32660
|
AtRLP22, receptor like protein 22, RLP22, receptor like protein 22 |
0.762 |
-0.128 |
| 29 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
0.761 |
0.007 |
| 30 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
0.760 |
0.114 |
| 31 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
0.756 |
-0.262 |
| 32 |
257021_at |
AT3G19710
|
BCAT4, branched-chain aminotransferase4 |
0.726 |
0.009 |
| 33 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.725 |
0.082 |
| 34 |
249955_at |
AT5G18840
|
[Major facilitator superfamily protein] |
0.723 |
0.023 |
| 35 |
264301_at |
AT1G78780
|
[pathogenesis-related family protein] |
0.721 |
0.038 |
| 36 |
256989_at |
AT3G28580
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.717 |
0.143 |
| 37 |
258941_at |
AT3G09940
|
ATMDAR3, ARABIDOPSIS THALIANA MONODEHYDROASCORBATE REDUCTASE 3, MDAR2, MONODEHYDROASCORBATE REDUCTASE 2, MDAR3, MONODEHYDROASCORBATE REDUCTASE 3, MDHAR, monodehydroascorbate reductase |
0.711 |
-0.177 |
| 38 |
246001_at |
AT5G20790
|
unknown |
0.703 |
0.390 |
| 39 |
251884_at |
AT3G54150
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.700 |
0.096 |
| 40 |
257774_at |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
0.698 |
0.132 |
| 41 |
251743_at |
AT3G55890
|
[Yippee family putative zinc-binding protein] |
0.697 |
-0.018 |
| 42 |
248729_at |
AT5G48010
|
AtTHAS1, Arabidopsis thaliana thalianol synthase 1, THAS, THALIANOL SYNTHASE, THAS1, thalianol synthase 1 |
0.696 |
0.018 |
| 43 |
261216_at |
AT1G33030
|
[O-methyltransferase family protein] |
0.695 |
0.241 |
| 44 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
0.693 |
0.089 |
| 45 |
249971_at |
AT5G19110
|
[Eukaryotic aspartyl protease family protein] |
0.688 |
-0.063 |
| 46 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
0.688 |
0.457 |
| 47 |
250702_at |
AT5G06730
|
[Peroxidase superfamily protein] |
0.686 |
0.118 |
| 48 |
254101_at |
AT4G25000
|
AMY1, alpha-amylase-like, ATAMY1 |
0.685 |
0.304 |
| 49 |
260706_at |
AT1G32350
|
AOX1D, alternative oxidase 1D |
0.676 |
0.161 |
| 50 |
256368_at |
AT1G66800
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.671 |
-0.035 |
| 51 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
0.668 |
0.240 |
| 52 |
266486_at |
AT2G47950
|
unknown |
0.664 |
0.081 |
| 53 |
257540_at |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
0.664 |
0.009 |
| 54 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.660 |
0.043 |
| 55 |
256321_at |
AT1G55020
|
ATLOX1, ARABIDOPSIS LIPOXYGENASE 1, LOX1, lipoxygenase 1 |
0.659 |
0.114 |
| 56 |
258064_at |
AT3G14680
|
CYP72A14, cytochrome P450, family 72, subfamily A, polypeptide 14 |
0.659 |
0.297 |
| 57 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
0.656 |
0.218 |
| 58 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
0.654 |
0.002 |
| 59 |
249567_at |
AT5G38020
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.650 |
-0.008 |
| 60 |
253667_at |
AT4G30170
|
[Peroxidase family protein] |
0.648 |
0.278 |
| 61 |
264893_at |
AT1G23140
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.648 |
0.280 |
| 62 |
245151_at |
AT2G47550
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.641 |
0.012 |
| 63 |
259925_at |
AT1G75040
|
PR-5, PR5, pathogenesis-related gene 5 |
0.640 |
0.134 |
| 64 |
257919_at |
AT3G23250
|
ATMYB15, MYB DOMAIN PROTEIN 15, ATY19, MYB15, myb domain protein 15 |
0.640 |
-0.107 |
| 65 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
0.639 |
0.005 |
| 66 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.638 |
0.186 |
| 67 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
0.635 |
0.043 |
| 68 |
252618_at |
AT3G45140
|
ATLOX2, ARABIODOPSIS THALIANA LIPOXYGENASE 2, LOX2, lipoxygenase 2 |
0.630 |
0.158 |
| 69 |
258957_at |
AT3G01420
|
ALPHA-DOX1, alpha-dioxygenase 1, DIOX1, DOX1, PADOX-1, plant alpha dioxygenase 1 |
0.630 |
0.198 |
| 70 |
248227_at |
AT5G53820
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.629 |
0.140 |
| 71 |
267391_at |
AT2G44480
|
BGLU17, beta glucosidase 17 |
0.628 |
0.217 |
| 72 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
0.627 |
-0.029 |
| 73 |
250646_at |
AT5G06720
|
ATPA2, peroxidase 2, AtPRX53, PA2, peroxidase 2, PRX53, peroxidase 53 |
0.626 |
0.143 |
| 74 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
0.624 |
0.125 |
| 75 |
253193_at |
AT4G35380
|
[SEC7-like guanine nucleotide exchange family protein] |
0.622 |
0.059 |
| 76 |
254247_at |
AT4G23260
|
CRK18, cysteine-rich RLK (RECEPTOR-like protein kinase) 18 |
0.621 |
0.081 |
| 77 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
0.621 |
0.578 |
| 78 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
0.620 |
0.232 |
| 79 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
0.618 |
0.046 |
| 80 |
246825_at |
AT5G26260
|
[TRAF-like family protein] |
0.613 |
-0.164 |
| 81 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
0.611 |
0.073 |
| 82 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
0.611 |
-0.104 |
| 83 |
255630_at |
AT4G00700
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
0.609 |
0.069 |
| 84 |
267207_at |
AT2G30840
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.608 |
0.073 |
| 85 |
256969_at |
AT3G21080
|
[ABC transporter-related] |
0.608 |
-0.025 |
| 86 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
0.605 |
0.280 |
| 87 |
264635_at |
AT1G65500
|
unknown |
0.599 |
0.041 |
| 88 |
260406_at |
AT1G69920
|
ATGSTU12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 12, GSTU12, glutathione S-transferase TAU 12 |
0.598 |
0.029 |
| 89 |
265341_at |
AT2G18360
|
[alpha/beta-Hydrolases superfamily protein] |
0.591 |
0.020 |
| 90 |
264729_at |
AT1G22990
|
HIPP22, heavy metal associated isoprenylated plant protein 22 |
0.587 |
-0.017 |
| 91 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.584 |
-0.001 |
| 92 |
249046_at |
AT5G44400
|
[FAD-binding Berberine family protein] |
0.584 |
0.055 |
| 93 |
248540_at |
AT5G50160
|
ATFRO8, FERRIC REDUCTION OXIDASE 8, FRO8, ferric reduction oxidase 8 |
0.569 |
-0.003 |
| 94 |
245436_at |
AT4G16620
|
UMAMIT8, Usually multiple acids move in and out Transporters 8 |
0.567 |
-0.046 |
| 95 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
0.562 |
-0.005 |
| 96 |
250474_at |
AT5G10230
|
ANN7, ANNEXIN 7, ANNAT7, annexin 7 |
0.557 |
-0.034 |
| 97 |
267459_at |
AT2G33850
|
unknown |
0.557 |
0.041 |
| 98 |
252488_at |
AT3G46700
|
[UDP-Glycosyltransferase superfamily protein] |
0.556 |
-0.000 |
| 99 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
0.555 |
-0.034 |
| 100 |
259743_at |
AT1G71140
|
[MATE efflux family protein] |
0.554 |
0.198 |
| 101 |
260541_at |
AT2G43530
|
[Scorpion toxin-like knottin superfamily protein] |
0.554 |
0.081 |
| 102 |
259640_at |
AT1G52400
|
ATBG1, A. THALIANA BETA-GLUCOSIDASE 1, BGL1, BETA-GLUCOSIDASE HOMOLOG 1, BGLU18, beta glucosidase 18 |
0.553 |
0.097 |
| 103 |
247105_at |
AT5G66630
|
DAR5, DA1-related protein 5 |
0.550 |
-0.050 |
| 104 |
248545_at |
AT5G50260
|
CEP1, cysteine endopeptidase 1 |
0.549 |
0.129 |
| 105 |
262378_at |
AT1G72830
|
ATHAP2C, HAP2C, NF-YA3, nuclear factor Y, subunit A3 |
0.548 |
0.132 |
| 106 |
250438_at |
AT5G10580
|
unknown |
0.548 |
0.002 |
| 107 |
259120_at |
AT3G02240
|
GLV4, GOLVEN 4, RGF7, root meristem growth factor 7 |
0.548 |
0.035 |
| 108 |
263475_at |
AT2G31945
|
unknown |
0.544 |
-0.082 |
| 109 |
249675_at |
AT5G35940
|
[Mannose-binding lectin superfamily protein] |
0.543 |
-0.000 |
| 110 |
260684_at |
AT1G17590
|
NF-YA8, nuclear factor Y, subunit A8 |
0.540 |
0.065 |
| 111 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
0.540 |
0.108 |
| 112 |
251986_at |
AT3G53310
|
[AP2/B3-like transcriptional factor family protein] |
0.539 |
-0.008 |
| 113 |
246197_at |
AT4G37010
|
CEN2, centrin 2 |
0.537 |
-0.025 |
| 114 |
258063_at |
AT3G14620
|
CYP72A8, cytochrome P450, family 72, subfamily A, polypeptide 8 |
0.536 |
0.216 |
| 115 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
0.535 |
-0.017 |
| 116 |
267226_at |
AT2G44010
|
unknown |
0.532 |
-0.003 |
| 117 |
260101_at |
AT1G73260
|
ATKTI1, ARABIDOPSIS THALIANA KUNITZ TRYPSIN INHIBITOR 1, KTI1, kunitz trypsin inhibitor 1 |
0.532 |
0.165 |
| 118 |
246682_at |
AT5G33290
|
XGD1, xylogalacturonan deficient 1 |
0.530 |
0.129 |
| 119 |
256877_at |
AT3G26470
|
[Powdery mildew resistance protein, RPW8 domain] |
0.529 |
0.173 |
| 120 |
247344_at |
AT5G63750
|
ARI13, ARIADNE 13, ATARI13, ARABIDOPSIS ARIADNE 13 |
0.526 |
-0.012 |
| 121 |
250365_at |
AT5G11410
|
[Protein kinase superfamily protein] |
0.525 |
0.059 |
| 122 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.524 |
0.144 |
| 123 |
254687_at |
AT4G13770
|
CYP83A1, cytochrome P450, family 83, subfamily A, polypeptide 1, REF2, REDUCED EPIDERMAL FLUORESCENCE 2 |
0.524 |
0.030 |
| 124 |
266712_at |
AT2G46750
|
AtGulLO2, GulLO2, L -gulono-1,4-lactone ( L -GulL) oxidase 2 |
0.523 |
-0.077 |
| 125 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
0.522 |
-0.013 |
| 126 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
0.520 |
0.060 |
| 127 |
265121_at |
AT1G62560
|
FMO GS-OX3, flavin-monooxygenase glucosinolate S-oxygenase 3 |
0.519 |
0.074 |
| 128 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
0.518 |
0.022 |
| 129 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
0.517 |
0.172 |
| 130 |
263565_at |
AT2G15390
|
atfut4, FUT4, fucosyltransferase 4 |
0.515 |
0.056 |
| 131 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
0.513 |
-0.014 |
| 132 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
0.511 |
0.006 |
| 133 |
252414_at |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
0.510 |
0.165 |
| 134 |
258890_at |
AT3G05690
|
ATHAP2B, HEME ACTIVATOR PROTEIN (YEAST) HOMOLOG 2B, HAP2B, HEME ACTIVATOR PROTEIN (YEAST) HOMOLOG 2B, NF-YA2, nuclear factor Y, subunit A2, UNE8, UNFERTILIZED EMBRYO SAC 8 |
0.510 |
0.062 |
| 135 |
252403_at |
AT3G48080
|
[alpha/beta-Hydrolases superfamily protein] |
0.509 |
-0.047 |
| 136 |
265014_at |
AT1G24430
|
[HXXXD-type acyl-transferase family protein] |
0.505 |
0.001 |
| 137 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.502 |
0.323 |
| 138 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
0.502 |
-0.003 |
| 139 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
0.502 |
0.062 |
| 140 |
263847_at |
AT2G36970
|
[UDP-Glycosyltransferase superfamily protein] |
0.502 |
0.262 |
| 141 |
248060_at |
AT5G55560
|
[Protein kinase superfamily protein] |
0.501 |
0.105 |
| 142 |
264887_at |
AT1G23120
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.500 |
-0.029 |
| 143 |
254447_at |
AT4G20860
|
[FAD-binding Berberine family protein] |
0.500 |
-0.089 |
| 144 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
0.496 |
0.057 |
| 145 |
259554_at |
AT1G21220
|
[copia-like retrotransposon family, has a 3.9e-26 P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
0.494 |
0.056 |
| 146 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
0.492 |
0.213 |
| 147 |
264680_at |
AT1G65510
|
unknown |
0.491 |
-0.115 |
| 148 |
259609_at |
AT1G52410
|
AtTSA1, TSA1, TSK-associating protein 1 |
0.489 |
0.132 |
| 149 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
0.487 |
0.190 |
| 150 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.485 |
-0.056 |
| 151 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
0.485 |
0.079 |
| 152 |
249576_at |
AT5G37690
|
[SGNH hydrolase-type esterase superfamily protein] |
0.484 |
0.117 |
| 153 |
249333_at |
AT5G40990
|
GLIP1, GDSL lipase 1 |
0.483 |
0.082 |
| 154 |
265796_at |
AT2G35730
|
[Heavy metal transport/detoxification superfamily protein ] |
0.482 |
0.025 |
| 155 |
252240_at |
AT3G50010
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.481 |
-0.010 |
| 156 |
248728_at |
AT5G48000
|
CYP708 A2, CYTOCHROME P450, FAMILY 708, SUBFAMILY A, POLYPEPTIDE 2, CYP708A2, cytochrome P450, family 708, subfamily A, polypeptide 2, THAH, THALIANOL HYDROXYLASE, THAH1, THALIANOL HYDROXYLASE 1 |
0.480 |
0.026 |
| 157 |
256878_at |
AT3G26460
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.479 |
-0.049 |
| 158 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
0.479 |
0.040 |
| 159 |
267169_at |
AT2G37540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.479 |
0.019 |
| 160 |
258596_at |
AT3G04510
|
LSH2, LIGHT SENSITIVE HYPOCOTYLS 2 |
0.478 |
0.127 |
| 161 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
0.477 |
-0.119 |
| 162 |
249454_at |
AT5G39520
|
unknown |
0.477 |
0.086 |
| 163 |
261242_at |
AT1G32960
|
ATSBT3.3, SBT3.3 |
0.476 |
0.101 |
| 164 |
252334_at |
AT3G48850
|
MPT2, mitochondrial phosphate transporter 2, PHT3;2, phosphate transporter 3;2 |
0.471 |
0.203 |
| 165 |
247225_at |
AT5G65090
|
BST1, BRISTLED 1, DER4, DEFORMED ROOT HAIRS 4, MRH3 |
0.471 |
0.069 |
| 166 |
255360_at |
AT4G03960
|
AtPFA-DSP4, PFA-DSP4, plant and fungi atypical dual-specificity phosphatase 4 |
0.468 |
0.218 |
| 167 |
247215_at |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
0.465 |
-0.014 |
| 168 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
0.463 |
0.105 |
| 169 |
260611_at |
AT2G43670
|
[Carbohydrate-binding X8 domain superfamily protein] |
0.461 |
0.004 |
| 170 |
253392_at |
AT4G32650
|
ATKC1, ARABIDOPSIS THALIANA K+ RECTIFYING CHANNEL 1, AtLKT1, A. thaliana low-K+-tolerant 1, KAT3, potassium channel in Arabidopsis thaliana 3, KC1 |
0.457 |
0.008 |
| 171 |
248539_at |
AT5G50130
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.456 |
-0.006 |
| 172 |
261033_at |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
0.456 |
0.053 |
| 173 |
260582_at |
AT2G47200
|
unknown |
0.455 |
0.027 |
| 174 |
261394_at |
AT1G79680
|
ATWAKL10, WAKL10, WALL ASSOCIATED KINASE (WAK)-LIKE 10 |
0.454 |
-0.077 |
| 175 |
254387_at |
AT4G21850
|
ATMSRB9, methionine sulfoxide reductase B9, MSRB9, methionine sulfoxide reductase B9 |
0.454 |
-0.115 |
| 176 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.453 |
0.598 |
| 177 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
0.452 |
0.149 |
| 178 |
257718_at |
AT3G18400
|
anac058, NAC domain containing protein 58, NAC058, NAC domain containing protein 58 |
0.451 |
-0.009 |
| 179 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
0.450 |
0.218 |
| 180 |
246368_at |
AT1G51890
|
[Leucine-rich repeat protein kinase family protein] |
0.449 |
-0.063 |
| 181 |
249188_at |
AT5G42830
|
[HXXXD-type acyl-transferase family protein] |
0.448 |
0.031 |
| 182 |
256774_at |
AT3G13760
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.448 |
-0.034 |
| 183 |
250724_at |
AT5G06330
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.448 |
0.103 |
| 184 |
260239_at |
AT1G74360
|
[Leucine-rich repeat protein kinase family protein] |
0.446 |
0.011 |
| 185 |
261449_at |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
0.445 |
0.008 |
| 186 |
260804_at |
AT1G78410
|
[VQ motif-containing protein] |
0.442 |
0.101 |
| 187 |
249527_at |
AT5G38710
|
[Methylenetetrahydrofolate reductase family protein] |
0.436 |
0.042 |
| 188 |
262940_at |
AT1G79520
|
[Cation efflux family protein] |
0.436 |
0.031 |
| 189 |
251479_at |
AT3G59700
|
ATHLECRK, lectin-receptor kinase, HLECRK, lectin-receptor kinase, LecRK-V.5, L-type lectin receptor kinase V.5, LecRK-V.5, LEGUME-LIKE LECTIN RECEPTOR KINASE-V.5, LECRK1, LECTIN-RECEPTOR KINASE 1 |
0.435 |
0.100 |
| 190 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
0.435 |
0.060 |
| 191 |
258203_at |
AT3G13950
|
unknown |
0.433 |
0.137 |
| 192 |
259033_at |
AT3G09405
|
[Pectinacetylesterase family protein] |
0.433 |
0.032 |
| 193 |
247794_at |
AT5G58670
|
ATPLC, ARABIDOPSIS THALIANA PHOSPHOLIPASE C, ATPLC1, phospholipase C1, PLC1, phospholipase C 1, PLC1, phospholipase C1 |
0.433 |
-0.028 |
| 194 |
256526_at |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
0.430 |
-0.029 |
| 195 |
254648_at |
AT4G18550
|
AtDSEL, Arabidopsis thaliana DAD1-like seeding establishment-related lipase, DSEL, DAD1-like seeding establishment-related lipase |
0.428 |
0.031 |
| 196 |
261240_at |
AT1G32940
|
ATSBT3.5, SBT3.5 |
0.427 |
0.050 |
| 197 |
254189_at |
AT4G24000
|
ATCSLG2, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G2, CSLG2, cellulose synthase like G2 |
0.426 |
0.292 |
| 198 |
265922_at |
AT2G18480
|
[Major facilitator superfamily protein] |
0.425 |
0.084 |
| 199 |
252076_at |
AT3G51660
|
[Tautomerase/MIF superfamily protein] |
0.425 |
-0.066 |
| 200 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
0.423 |
0.195 |
| 201 |
257373_at |
AT2G43140
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.423 |
0.123 |
| 202 |
252966_at |
AT4G38870
|
[F-box and associated interaction domains-containing protein] |
0.422 |
0.036 |
| 203 |
257512_at |
AT1G35250
|
ALT2, acyl-lipid thioesterase 2 |
0.419 |
-0.031 |
| 204 |
249777_at |
AT5G24210
|
[alpha/beta-Hydrolases superfamily protein] |
0.418 |
0.043 |
| 205 |
261899_at |
AT1G80820
|
ATCCR2, CCR2, cinnamoyl coa reductase |
0.417 |
-0.011 |
| 206 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
0.417 |
-0.005 |
| 207 |
247196_at |
AT5G65510
|
AIL7, AINTEGUMENTA-like 7, PLT7, PLETHORA 7 |
0.417 |
0.021 |
| 208 |
252214_at |
AT3G50260
|
ATERF#011, CEJ1, cooperatively regulated by ethylene and jasmonate 1, DEAR1, DREB AND EAR MOTIF PROTEIN 1 |
0.416 |
0.085 |
| 209 |
252763_at |
AT3G42725
|
[Putative membrane lipoprotein] |
0.416 |
0.000 |
| 210 |
251304_at |
AT3G61990
|
OMTF3, O-MTase family 3 protein |
0.415 |
0.232 |
| 211 |
265769_at |
AT2G48090
|
unknown |
0.415 |
0.073 |
| 212 |
257074_at |
AT3G19660
|
unknown |
0.415 |
0.077 |
| 213 |
250669_at |
AT5G06870
|
ATPGIP2, ARABIDOPSIS POLYGALACTURONASE INHIBITING PROTEIN 2, PGIP2, polygalacturonase inhibiting protein 2 |
0.415 |
-0.020 |
| 214 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.414 |
0.546 |
| 215 |
262314_at |
AT1G70810
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.414 |
0.157 |
| 216 |
251466_at |
AT3G59340
|
unknown |
0.413 |
-0.045 |
| 217 |
258787_at |
AT3G11840
|
PUB24, plant U-box 24 |
0.412 |
0.020 |
| 218 |
255059_at |
AT4G09420
|
[Disease resistance protein (TIR-NBS class)] |
0.411 |
-0.078 |
| 219 |
250483_at |
AT5G10300
|
AtHNL, ATMES5, ARABIDOPSIS THALIANA METHYL ESTERASE 5, HNL, HYDROXYNITRILE LYASE, MES5, methyl esterase 5 |
0.411 |
0.108 |
| 220 |
261913_at |
AT1G65860
|
FMO GS-OX1, flavin-monooxygenase glucosinolate S-oxygenase 1 |
0.411 |
0.031 |
| 221 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
0.411 |
0.072 |
| 222 |
258570_at |
AT3G04530
|
ATPPCK2, PEPCK2, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 2, PPCK2, phosphoenolpyruvate carboxylase kinase 2 |
0.408 |
0.402 |
| 223 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
0.408 |
0.241 |
| 224 |
256750_at |
AT3G27150
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.407 |
0.060 |
| 225 |
267411_at |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
0.404 |
-0.089 |
| 226 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.404 |
0.196 |
| 227 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
0.404 |
-0.153 |
| 228 |
249814_at |
AT5G23840
|
[MD-2-related lipid recognition domain-containing protein] |
0.403 |
-0.032 |
| 229 |
249061_at |
AT5G44550
|
[Uncharacterised protein family (UPF0497)] |
0.403 |
-0.014 |
| 230 |
264960_at |
AT1G76930
|
ATEXT1, EXTENSIN 1, ATEXT4, extensin 4, EXT1, extensin 1, EXT4, extensin 4, ORG5, OBP3-RESPONSIVE GENE 5 |
0.403 |
0.013 |
| 231 |
265025_at |
AT1G24575
|
unknown |
0.403 |
0.065 |
| 232 |
261443_at |
AT1G28480
|
GRX480, roxy19 |
0.403 |
0.005 |
| 233 |
266658_at |
AT2G25735
|
unknown |
0.402 |
-0.055 |
| 234 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
0.402 |
-0.011 |
| 235 |
259653_at |
AT1G55240
|
unknown |
0.402 |
-0.027 |
| 236 |
251766_at |
AT3G55910
|
unknown |
0.401 |
0.136 |
| 237 |
252921_at |
AT4G39030
|
EDS5, ENHANCED DISEASE SUSCEPTIBILITY 5, SCORD3, susceptible to coronatine-deficient Pst DC3000 3, SID1, SALICYLIC ACID INDUCTION DEFICIENT 1 |
0.400 |
0.019 |
| 238 |
264815_at |
AT1G03620
|
[ELMO/CED-12 family protein] |
0.400 |
-0.081 |
| 239 |
264616_at |
AT2G17740
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.400 |
-0.158 |
| 240 |
260092_at |
AT1G73280
|
scpl3, serine carboxypeptidase-like 3 |
0.400 |
0.001 |
| 241 |
261199_at |
AT1G12950
|
RSH2, root hair specific 2 |
0.399 |
0.094 |
| 242 |
249896_at |
AT5G22530
|
unknown |
0.399 |
0.026 |
| 243 |
264305_at |
AT1G78815
|
LSH7, LIGHT SENSITIVE HYPOCOTYLS 7 |
0.398 |
0.067 |
| 244 |
259553_x_at |
AT1G21310
|
ATEXT3, extensin 3, EXT3, extensin 3, RSH, ROOT-SHOOT-HYPOCOTYL DEFECTIVE |
0.398 |
0.279 |
| 245 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
-1.283 |
-0.187 |
| 246 |
254122_at |
AT4G24510
|
CER2, ECERIFERUM 2, VC-2, VC2 |
-1.188 |
-0.055 |
| 247 |
259104_at |
AT3G02170
|
LNG2, LONGIFOLIA2, TRM1, TON1 Recruiting Motif 1 |
-1.136 |
-0.100 |
| 248 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-1.101 |
-0.256 |
| 249 |
248921_at |
AT5G45950
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-1.080 |
-0.082 |
| 250 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
-1.080 |
-0.081 |
| 251 |
245336_at |
AT4G16515
|
CLEL 6, CLE-like 6, GLV1, GOLVEN 1, RGF6, root meristem growth factor 6 |
-1.054 |
-0.168 |
| 252 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-1.052 |
-0.165 |
| 253 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
-1.050 |
-0.137 |
| 254 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-1.045 |
-0.294 |
| 255 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-1.032 |
-0.179 |
| 256 |
253255_at |
AT4G34760
|
SAUR50, SMALL AUXIN UPREGULATED RNA 50 |
-1.031 |
-0.174 |
| 257 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-1.025 |
-0.337 |
| 258 |
265400_at |
AT2G10940
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-1.016 |
-0.049 |
| 259 |
257701_at |
AT3G12710
|
[DNA glycosylase superfamily protein] |
-1.016 |
0.028 |
| 260 |
254041_at |
AT4G25830
|
[Uncharacterised protein family (UPF0497)] |
-0.972 |
-0.034 |
| 261 |
261226_at |
AT1G20190
|
ATEXP11, ATEXPA11, expansin 11, ATHEXP ALPHA 1.14, EXP11, EXPANSIN 11, EXPA11, expansin 11 |
-0.966 |
-0.066 |
| 262 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
-0.961 |
-0.253 |
| 263 |
246562_at |
AT5G15580
|
LNG1, LONGIFOLIA1, TRM2, TON1 Recruiting Motif 2 |
-0.952 |
-0.075 |
| 264 |
259786_at |
AT1G29660
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.951 |
-0.027 |
| 265 |
252765_at |
AT3G42800
|
unknown |
-0.938 |
-0.187 |
| 266 |
267545_at |
AT2G32690
|
ATGRP23, GLYCINE-RICH PROTEIN 23, GRP23, glycine-rich protein 23 |
-0.937 |
-0.139 |
| 267 |
246275_at |
AT4G36540
|
BEE2, BR enhanced expression 2 |
-0.935 |
-0.115 |
| 268 |
250366_at |
AT5G11420
|
unknown |
-0.926 |
-0.111 |
| 269 |
247880_at |
AT5G57780
|
P1R1, P1R1 |
-0.911 |
-0.127 |
| 270 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
-0.900 |
0.039 |
| 271 |
248509_at |
AT5G50335
|
unknown |
-0.897 |
0.172 |
| 272 |
254110_at |
AT4G25260
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.893 |
-0.143 |
| 273 |
258003_at |
AT3G29030
|
ATEXP5, ARABIDOPSIS THALIANA EXPANSIN 5, ATEXPA5, ARABIDOPSIS THALIANA EXPANSIN A5, ATHEXP ALPHA 1.4, EXP5, EXPANSIN 5, EXPA5, expansin A5 |
-0.879 |
-0.029 |
| 274 |
247252_at |
AT5G64770
|
CLEL 9, CLE-like 9, GLV2, GOLVEN 2, RGF9, root meristem growth factor 9 |
-0.877 |
-0.135 |
| 275 |
249894_at |
AT5G22580
|
[Stress responsive A/B Barrel Domain] |
-0.877 |
0.039 |
| 276 |
259375_at |
AT3G16370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.872 |
-0.064 |
| 277 |
265806_at |
AT2G18010
|
SAUR10, SMALL AUXIN UPREGULATED RNA 10 |
-0.862 |
-0.088 |
| 278 |
250936_at |
AT5G03120
|
unknown |
-0.861 |
-0.228 |
| 279 |
254609_at |
AT4G18970
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.852 |
-0.014 |
| 280 |
263765_at |
AT2G21540
|
ATSFH3, SEC14-LIKE 3, SFH3, SEC14-like 3 |
-0.847 |
-0.197 |
| 281 |
258552_at |
AT3G07010
|
[Pectin lyase-like superfamily protein] |
-0.841 |
-0.080 |
| 282 |
253207_at |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
-0.839 |
0.071 |
| 283 |
254685_at |
AT4G13790
|
SAUR25, SMALL AUXIN UPREGULATED RNA 25 |
-0.838 |
0.015 |
| 284 |
254327_at |
AT4G22490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.838 |
-0.035 |
| 285 |
267158_at |
AT2G37640
|
ATEXP3, EXPANSIN 3, ATEXPA3, ARABIDOPSIS THALIANA EXPANSIN A3, ATHEXP ALPHA 1.9, EXP3 |
-0.835 |
0.017 |
| 286 |
247377_at |
AT5G63180
|
[Pectin lyase-like superfamily protein] |
-0.831 |
-0.001 |
| 287 |
259788_at |
AT1G29670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.827 |
-0.072 |
| 288 |
247288_at |
AT5G64330
|
JK218, NPH3, NON-PHOTOTROPIC HYPOCOTYL 3, RPT3, ROOT PHOTOTROPISM 3 |
-0.813 |
-0.023 |
| 289 |
262730_at |
AT1G16390
|
ATOCT3, organic cation/carnitine transporter 3, OCT3, organic cation/carnitine transporter 3 |
-0.813 |
-0.097 |
| 290 |
252296_at |
AT3G48970
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.807 |
-0.096 |
| 291 |
250892_at |
AT5G03760
|
ATCSLA09, ATCSLA9, CSLA09, CSLA9, CELLULOSE SYNTHASE LIKE A9, RAT4, RESISTANT TO AGROBACTERIUM TRANSFORMATION 4 |
-0.807 |
-0.045 |
| 292 |
255694_at |
AT4G00050
|
UNE10, unfertilized embryo sac 10 |
-0.806 |
0.060 |
| 293 |
249008_at |
AT5G44680
|
[DNA glycosylase superfamily protein] |
-0.789 |
-0.154 |
| 294 |
245318_at |
AT4G16980
|
[arabinogalactan-protein family] |
-0.789 |
-0.105 |
| 295 |
260941_at |
AT1G44970
|
[Peroxidase superfamily protein] |
-0.785 |
0.334 |
| 296 |
261772_at |
AT1G76240
|
unknown |
-0.783 |
-0.075 |
| 297 |
259785_at |
AT1G29490
|
SAUR68, SMALL AUXIN UPREGULATED 68 |
-0.782 |
-0.005 |
| 298 |
265342_at |
AT2G18300
|
HBI1, HOMOLOG OF BEE2 INTERACTING WITH IBH 1 |
-0.778 |
-0.128 |
| 299 |
252970_at |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
-0.776 |
-0.082 |
| 300 |
252971_at |
AT4G38770
|
ATPRP4, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 4, PRP4, proline-rich protein 4 |
-0.776 |
-0.030 |
| 301 |
252943_at |
AT4G39330
|
ATCAD9, CAD9, cinnamyl alcohol dehydrogenase 9 |
-0.775 |
0.027 |
| 302 |
254572_at |
AT4G19380
|
[Long-chain fatty alcohol dehydrogenase family protein] |
-0.773 |
-0.242 |
| 303 |
265645_at |
AT2G27370
|
CASP3, Casparian strip membrane domain protein 3 |
-0.766 |
-0.011 |
| 304 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
-0.760 |
-0.186 |
| 305 |
261745_at |
AT1G08500
|
AtENODL18, ENODL18, early nodulin-like protein 18 |
-0.755 |
-0.003 |
| 306 |
266356_at |
AT2G32300
|
UCC1, uclacyanin 1 |
-0.754 |
-0.000 |
| 307 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
-0.752 |
0.034 |
| 308 |
261825_at |
AT1G11545
|
XTH8, xyloglucan endotransglucosylase/hydrolase 8 |
-0.750 |
-0.103 |
| 309 |
250277_at |
AT5G12940
|
[Leucine-rich repeat (LRR) family protein] |
-0.747 |
-0.000 |
| 310 |
259549_at |
AT1G35290
|
ALT1, acyl-lipid thioesterase 1 |
-0.745 |
0.082 |
| 311 |
258745_at |
AT3G05920
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.743 |
0.009 |
| 312 |
259839_at |
AT1G52190
|
AtNPF1.2, NPF1.2, NRT1/ PTR family 1.2, NRT1.11, nitrate transporter 1.11 |
-0.741 |
-0.045 |
| 313 |
259660_at |
AT1G55260
|
LTPG6, glycosylphosphatidylinositol-anchored lipid protein transfer 6 |
-0.740 |
-0.034 |
| 314 |
250657_at |
AT5G07000
|
ATST2B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2B, ST2B, sulfotransferase 2B |
-0.739 |
0.118 |
| 315 |
259332_at |
AT3G03830
|
SAUR28, SMALL AUXIN UP RNA 28 |
-0.732 |
-0.094 |
| 316 |
247085_at |
AT5G66310
|
[ATP binding microtubule motor family protein] |
-0.731 |
-0.042 |
| 317 |
263979_at |
AT2G42840
|
PDF1, protodermal factor 1 |
-0.730 |
-0.085 |
| 318 |
266460_at |
AT2G47930
|
AGP26, arabinogalactan protein 26, ATAGP26, ARABIDOPSIS THALIANA ARABINOGALACTAN PROTEIN 26 |
-0.723 |
-0.180 |
| 319 |
264007_at |
AT2G21140
|
ATPRP2, proline-rich protein 2, PRP2, proline-rich protein 2 |
-0.718 |
0.028 |
| 320 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
-0.715 |
-0.180 |
| 321 |
259398_at |
AT1G17700
|
PRA1.F1, prenylated RAB acceptor 1.F1 |
-0.712 |
-0.102 |
| 322 |
250891_at |
AT5G04530
|
KCS19, 3-ketoacyl-CoA synthase 19 |
-0.707 |
0.018 |
| 323 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
-0.703 |
0.017 |
| 324 |
259624_at |
AT1G43020
|
unknown |
-0.699 |
-0.043 |
| 325 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
-0.692 |
-0.236 |
| 326 |
261480_at |
AT1G14280
|
PKS2, phytochrome kinase substrate 2 |
-0.692 |
-0.134 |
| 327 |
264901_at |
AT1G23090
|
AST91, sulfate transporter 91, SULTR3;3 |
-0.687 |
0.090 |
| 328 |
249227_at |
AT5G42180
|
PER64, peroxidase 64 |
-0.683 |
0.209 |
| 329 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
-0.679 |
0.024 |
| 330 |
262978_at |
AT1G75780
|
TUB1, tubulin beta-1 chain |
-0.679 |
-0.001 |
| 331 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
-0.678 |
-0.205 |
| 332 |
255538_at |
AT4G01680
|
AtMYB55, myb domain protein 55, MYB55, myb domain protein 55 |
-0.676 |
-0.002 |
| 333 |
261641_at |
AT1G27670
|
unknown |
-0.670 |
-0.130 |
| 334 |
263809_at |
AT2G04570
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.669 |
0.088 |
| 335 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
-0.666 |
-0.271 |
| 336 |
258367_at |
AT3G14370
|
WAG2 |
-0.662 |
0.193 |
| 337 |
251183_at |
AT3G62630
|
unknown |
-0.660 |
-0.034 |
| 338 |
258589_at |
AT3G04290
|
ATLTL1, LTL1, Li-tolerant lipase 1 |
-0.660 |
0.021 |
| 339 |
248681_at |
AT5G48900
|
[Pectin lyase-like superfamily protein] |
-0.659 |
-0.067 |
| 340 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-0.658 |
-0.187 |
| 341 |
252972_at |
AT4G38840
|
SAUR14, SMALL AUXIN UPREGULATED RNA 14 |
-0.658 |
-0.075 |
| 342 |
262682_at |
AT1G75900
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.655 |
-0.036 |
| 343 |
248028_at |
AT5G55620
|
unknown |
-0.649 |
0.016 |
| 344 |
245696_at |
AT5G04190
|
PKS4, phytochrome kinase substrate 4 |
-0.648 |
-0.158 |
| 345 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
-0.647 |
0.100 |
| 346 |
259161_at |
AT3G01500
|
ATBCA1, BETA CARBONIC ANHYDRASE 1, ATSABP3, ARABIDOPSIS THALIANA SALICYLIC ACID-BINDING PROTEIN 3, CA1, carbonic anhydrase 1, SABP3, SALICYLIC ACID-BINDING PROTEIN 3 |
-0.647 |
-0.058 |
| 347 |
267013_at |
AT2G39180
|
ATCRR2, CCR2, CRINKLY4 related 2 |
-0.646 |
-0.044 |
| 348 |
254718_at |
AT4G13580
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.639 |
0.041 |
| 349 |
265246_at |
AT2G43050
|
ATPMEPCRD |
-0.633 |
-0.193 |
| 350 |
249755_at |
AT5G24580
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.630 |
0.004 |
| 351 |
261975_at |
AT1G64640
|
AtENODL8, ENODL8, early nodulin-like protein 8 |
-0.630 |
-0.118 |
| 352 |
264021_at |
AT2G21200
|
SAUR7, SMALL AUXIN UPREGULATED RNA 7 |
-0.626 |
-0.154 |
| 353 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.623 |
-0.027 |
| 354 |
254221_at |
AT4G23820
|
[Pectin lyase-like superfamily protein] |
-0.618 |
-0.116 |
| 355 |
246908_at |
AT5G25610
|
ATRD22, RD22, RESPONSIVE TO DESSICATION 22 |
-0.616 |
0.027 |
| 356 |
258742_at |
AT3G05800
|
AIF1, AtBS1(activation-tagged BRI1 suppressor 1)-interacting factor 1 |
-0.616 |
-0.079 |
| 357 |
250717_at |
AT5G06200
|
CASP4, Casparian strip membrane domain protein 4 |
-0.613 |
-0.029 |
| 358 |
263664_at |
AT1G04250
|
AXR3, AUXIN RESISTANT 3, IAA17, indole-3-acetic acid inducible 17 |
-0.609 |
0.009 |
| 359 |
266805_at |
AT2G30010
|
TBL45, TRICHOME BIREFRINGENCE-LIKE 45 |
-0.606 |
-0.098 |
| 360 |
253579_at |
AT4G30610
|
BRS1, BRI1 SUPPRESSOR 1, SCPL24, SERINE CARBOXYPEPTIDASE 24 PRECURSOR |
-0.605 |
-0.100 |
| 361 |
256237_at |
AT3G12610
|
DRT100, DNA-DAMAGE REPAIR/TOLERATION 100 |
-0.603 |
-0.028 |
| 362 |
266688_at |
AT2G19660
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.602 |
-0.138 |
| 363 |
246919_at |
AT5G25460
|
DGR2, DUF642 L-GalL responsive gene 2 |
-0.601 |
-0.037 |
| 364 |
263284_at |
AT2G36100
|
CASP1, Casparian strip membrane domain protein 1 |
-0.600 |
0.067 |
| 365 |
261949_at |
AT1G64670
|
BDG1, BODYGUARD1, CED1, 9-CIS EPOXYCAROTENOID DIOXYGENASE DEFECTIVE 1 |
-0.599 |
0.014 |
| 366 |
250108_at |
AT5G15150
|
ATHB-3, homeobox 3, ATHB3, HAT7, HOMEOBOX FROM ARABIDOPSIS THALIANA 7, HB-3, homeobox 3 |
-0.598 |
-0.042 |
| 367 |
259291_at |
AT3G11550
|
CASP2, Casparian strip membrane domain protein 2 |
-0.596 |
0.007 |
| 368 |
261203_at |
AT1G12845
|
unknown |
-0.594 |
-0.042 |
| 369 |
257297_at |
AT3G28040
|
[Leucine-rich receptor-like protein kinase family protein] |
-0.588 |
-0.071 |
| 370 |
264577_at |
AT1G05260
|
RCI3, RARE COLD INDUCIBLE GENE 3, RCI3A |
-0.588 |
0.185 |
| 371 |
263437_at |
AT2G28670
|
ESB1, ENHANCED SUBERIN 1 |
-0.588 |
0.040 |
| 372 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-0.587 |
-0.012 |
| 373 |
248252_at |
AT5G53250
|
AGP22, arabinogalactan protein 22, ATAGP22, ARABINOGALACTAN PROTEIN 22 |
-0.584 |
-0.304 |
| 374 |
250437_at |
AT5G10430
|
AGP4, arabinogalactan protein 4, ATAGP4 |
-0.583 |
-0.072 |
| 375 |
249887_at |
AT5G22310
|
unknown |
-0.581 |
0.066 |
| 376 |
246097_at |
AT5G20270
|
HHP1, heptahelical transmembrane protein1 |
-0.580 |
-0.054 |
| 377 |
267141_at |
AT2G38090
|
[Duplicated homeodomain-like superfamily protein] |
-0.579 |
0.058 |
| 378 |
267481_at |
AT2G02780
|
Leucine-rich repeat protein kinase family protein |
-0.577 |
-0.079 |
| 379 |
250165_at |
AT5G15290
|
CASP5, Casparian strip membrane domain protein 5 |
-0.575 |
-0.026 |
| 380 |
256743_at |
AT3G29370
|
P1R3, P1R3 |
-0.574 |
-0.053 |
| 381 |
248623_at |
AT5G49170
|
unknown |
-0.572 |
0.033 |
| 382 |
258145_at |
AT3G18200
|
UMAMIT4, Usually multiple acids move in and out Transporters 4 |
-0.570 |
-0.094 |
| 383 |
250982_at |
AT5G03150
|
JKD, JACKDAW |
-0.569 |
-0.020 |
| 384 |
246002_at |
AT5G20740
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.566 |
-0.022 |
| 385 |
247020_at |
AT5G67020
|
unknown |
-0.565 |
0.038 |
| 386 |
266978_at |
AT2G39430
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.565 |
-0.039 |
| 387 |
266320_at |
AT2G46640
|
TAC1, Tiller Angle Control 1 |
-0.557 |
0.132 |
| 388 |
261509_at |
AT1G71740
|
unknown |
-0.551 |
0.013 |
| 389 |
266613_at |
AT2G14900
|
[Gibberellin-regulated family protein] |
-0.548 |
-0.045 |
| 390 |
258857_at |
AT3G02110
|
scpl25, serine carboxypeptidase-like 25 |
-0.544 |
-0.023 |
| 391 |
252711_at |
AT3G43720
|
LTPG2, glycosylphosphatidylinositol-anchored lipid protein transfer 2 |
-0.544 |
-0.053 |
| 392 |
258675_at |
AT3G08770
|
LTP6, lipid transfer protein 6 |
-0.542 |
0.084 |
| 393 |
267376_at |
AT2G26330
|
ER, ERECTA, QRP1, QUANTITATIVE RESISTANCE TO PLECTOSPHAERELLA 1 |
-0.538 |
-0.077 |
| 394 |
250734_at |
AT5G06270
|
unknown |
-0.536 |
-0.107 |
| 395 |
263431_at |
AT2G22170
|
PLAT2, PLAT domain protein 2 |
-0.536 |
-0.006 |
| 396 |
258528_at |
AT3G06770
|
[Pectin lyase-like superfamily protein] |
-0.532 |
-0.144 |
| 397 |
247352_at |
AT5G63650
|
SNRK2-5, SNRK2.5, SNF1-related protein kinase 2.5, SRK2H, SNF1-RELATED PROTEIN KINASE 2H |
-0.531 |
-0.029 |
| 398 |
252115_at |
AT3G51600
|
LTP5, lipid transfer protein 5 |
-0.530 |
0.027 |
| 399 |
248371_at |
AT5G51810
|
AT2353, ATGA20OX2, GA20OX2, gibberellin 20 oxidase 2 |
-0.530 |
-0.100 |
| 400 |
261981_at |
AT1G33811
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.518 |
-0.038 |
| 401 |
257203_at |
AT3G23730
|
XTH16, xyloglucan endotransglucosylase/hydrolase 16 |
-0.518 |
-0.065 |
| 402 |
260806_at |
AT1G78260
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.517 |
-0.121 |
| 403 |
245574_at |
AT4G14750
|
IQD19, IQ-domain 19 |
-0.516 |
-0.092 |
| 404 |
250565_at |
AT5G08000
|
E13L3, glucan endo-1,3-beta-glucosidase-like protein 3, PDCB2, PLASMODESMATA CALLOSE-BINDING PROTEIN 2 |
-0.507 |
-0.045 |
| 405 |
258751_at |
AT3G05890
|
RCI2B, RARE-COLD-INDUCIBLE 2B |
-0.507 |
-0.225 |
| 406 |
249010_at |
AT5G44580
|
unknown |
-0.500 |
0.004 |
| 407 |
255774_at |
AT1G18620
|
TRM3, TON1 Recruiting Motif 3 |
-0.497 |
-0.167 |
| 408 |
261917_at |
AT1G65920
|
[Regulator of chromosome condensation (RCC1) family with FYVE zinc finger domain] |
-0.496 |
0.034 |
| 409 |
253754_at |
AT4G29020
|
[glycine-rich protein] |
-0.495 |
-0.027 |
| 410 |
247170_at |
AT5G65530
|
[Protein kinase superfamily protein] |
-0.494 |
0.020 |
| 411 |
249588_at |
AT5G37790
|
[Protein kinase superfamily protein] |
-0.493 |
-0.100 |
| 412 |
250653_at |
AT5G06930
|
[LOCATED IN: chloroplast] |
-0.493 |
-0.145 |
| 413 |
258764_at |
AT3G10720
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.491 |
-0.124 |
| 414 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
-0.490 |
-0.247 |
| 415 |
256415_at |
AT3G11210
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.489 |
0.110 |
| 416 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.488 |
-0.270 |
| 417 |
252863_at |
AT4G39800
|
ATIPS1, INOSITOL 3-PHOSPHATE SYNTHASE 1, ATMIPS1, MYO-INOSITOL-1-PHOSPHATE SYNTHASE 1, MI-1-P SYNTHASE, MIPS1, D-myo-Inositol 3-Phosphate Synthase 1, MIPS1, myo-inositol-1-phosphate synthase 1 |
-0.486 |
0.068 |
| 418 |
247362_at |
AT5G63140
|
ATPAP29, purple acid phosphatase 29, PAP29, PAP29, purple acid phosphatase 29 |
-0.485 |
-0.008 |
| 419 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
-0.484 |
-0.045 |
| 420 |
265877_at |
AT2G42380
|
ATBZIP34, BZIP34 |
-0.484 |
-0.182 |
| 421 |
258091_at |
AT3G14560
|
unknown |
-0.483 |
0.052 |
| 422 |
255854_at |
AT1G67050
|
unknown |
-0.483 |
-0.139 |
| 423 |
249278_at |
AT5G41900
|
[alpha/beta-Hydrolases superfamily protein] |
-0.482 |
-0.016 |
| 424 |
256452_at |
AT1G75240
|
AtHB33, homeobox protein 33, HB33, homeobox protein 33, ZHD5, zinc-finger homeodomain 5 |
-0.482 |
-0.030 |
| 425 |
252255_at |
AT3G49220
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.481 |
0.004 |
| 426 |
252412_at |
AT3G47295
|
unknown |
-0.475 |
-0.199 |
| 427 |
260483_at |
AT1G11000
|
ATMLO4, MILDEW RESISTANCE LOCUS O 4, MLO4, MILDEW RESISTANCE LOCUS O 4 |
-0.475 |
-0.011 |
| 428 |
258919_at |
AT3G10525
|
LGO, LOSS OF GIANT CELLS FROM ORGANS, SMR1, SIAMESE RELATED 1 |
-0.474 |
-0.102 |
| 429 |
264016_at |
AT2G21220
|
SAUR12, SMALL AUXIN UPREGULATED RNA 12 |
-0.472 |
0.102 |
| 430 |
255528_at |
AT4G02090
|
unknown |
-0.470 |
0.018 |
| 431 |
255642_at |
AT4G00820
|
iqd17, IQ-domain 17 |
-0.470 |
-0.073 |
| 432 |
250261_at |
AT5G13400
|
[Major facilitator superfamily protein] |
-0.470 |
-0.104 |
| 433 |
261327_at |
AT1G44830
|
[Integrase-type DNA-binding superfamily protein] |
-0.469 |
0.177 |
| 434 |
266392_at |
AT2G41280
|
ATM10, M10 |
-0.468 |
-0.054 |
| 435 |
255856_at |
AT1G66940
|
[protein kinase-related] |
-0.462 |
-0.007 |
| 436 |
254809_at |
AT4G12410
|
SAUR35, SMALL AUXIN UPREGULATED RNA 35 |
-0.460 |
0.085 |
| 437 |
260982_at |
AT1G53520
|
AtFAP3, FAP3, fatty-acid-binding protein 3 |
-0.459 |
-0.007 |
| 438 |
251330_at |
AT3G61550
|
[RING/U-box superfamily protein] |
-0.457 |
-0.111 |
| 439 |
247189_at |
AT5G65390
|
AGP7, arabinogalactan protein 7 |
-0.448 |
-0.129 |
| 440 |
256114_at |
AT1G16850
|
unknown |
-0.447 |
-0.023 |
| 441 |
255549_at |
AT4G01950
|
ATGPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3, GPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3 |
-0.447 |
-0.069 |
| 442 |
247214_at |
AT5G64850
|
unknown |
-0.446 |
-0.101 |
| 443 |
256066_at |
AT1G06980
|
unknown |
-0.446 |
-0.430 |
| 444 |
255732_at |
AT1G25450
|
CER60, ECERIFERUM 60, KCS5, 3-ketoacyl-CoA synthase 5 |
-0.444 |
-0.002 |
| 445 |
249847_at |
AT5G23210
|
SCPL34, serine carboxypeptidase-like 34 |
-0.442 |
-0.013 |
| 446 |
262376_at |
AT1G72970
|
EDA17, embryo sac development arrest 17, HTH, HOTHEAD |
-0.442 |
-0.033 |
| 447 |
250189_at |
AT5G14410
|
unknown |
-0.442 |
-0.172 |
| 448 |
257547_at |
AT3G13000
|
unknown |
-0.441 |
-0.098 |
| 449 |
246371_at |
AT1G51940
|
LYK3, LysM-containing receptor-like kinase 3 |
-0.440 |
-0.036 |
| 450 |
254737_at |
AT4G13840
|
CER26, ECERIFERUM 26 |
-0.440 |
-0.011 |
| 451 |
264902_at |
AT1G23060
|
MDP40, MICROTUBULE DESTABILIZING PROTEIN 40 |
-0.436 |
0.126 |
| 452 |
261292_at |
AT1G36940
|
unknown |
-0.435 |
-0.152 |
| 453 |
261594_at |
AT1G33240
|
AT-GTL1, GT2-LIKE 1, ATGTL1, GTL1, GT-2-like 1 |
-0.434 |
-0.145 |
| 454 |
249063_at |
AT5G44110
|
ABCI21, ATP-binding cassette A21, ATNAP2, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN, ATPOP1, POP1 |
-0.433 |
0.032 |
| 455 |
248208_at |
AT5G53980
|
ATHB52, homeobox protein 52, HB52, homeobox protein 52 |
-0.432 |
0.026 |
| 456 |
261308_at |
AT1G48480
|
RKL1, receptor-like kinase 1 |
-0.432 |
-0.054 |
| 457 |
250327_at |
AT5G12050
|
unknown |
-0.428 |
-0.244 |
| 458 |
252143_at |
AT3G51150
|
[ATP binding microtubule motor family protein] |
-0.425 |
-0.040 |
| 459 |
265249_at |
AT2G01940
|
ATIDD15, ARABIDOPSIS THALIANA INDETERMINATE(ID)-DOMAIN 15, SGR5, SHOOT GRAVITROPISM 5 |
-0.424 |
-0.112 |
| 460 |
246063_at |
AT5G19340
|
unknown |
-0.421 |
0.020 |
| 461 |
265688_at |
AT2G24300
|
[Calmodulin-binding protein] |
-0.421 |
-0.117 |
| 462 |
261819_at |
AT1G11410
|
[S-locus lectin protein kinase family protein] |
-0.420 |
-0.067 |
| 463 |
264378_at |
AT2G25220
|
[Protein kinase superfamily protein] |
-0.419 |
-0.042 |
| 464 |
245657_at |
AT1G56720
|
[Protein kinase superfamily protein] |
-0.418 |
-0.175 |
| 465 |
257137_at |
AT3G28860
|
ABCB19, ATP-binding cassette B19, ATABCB19, Arabidopsis thaliana ATP-binding cassette B19, ATMDR1, ATMDR11, MULTIDRUG RESISTANCE PROTEIN 11, ATPGP19, MDR1, MDR11, PGP19, P-GLYCOPROTEIN 19 |
-0.418 |
-0.103 |
| 466 |
252911_at |
AT4G39510
|
CYP96A12, cytochrome P450, family 96, subfamily A, polypeptide 12 |
-0.418 |
0.074 |
| 467 |
254034_at |
AT4G25960
|
ABCB2, ATP-binding cassette B2, PGP2, P-glycoprotein 2 |
-0.418 |
-0.130 |
| 468 |
247871_at |
AT5G57530
|
AtXTH12, XTH12, xyloglucan endotransglucosylase/hydrolase 12 |
-0.413 |
0.031 |
| 469 |
263218_at |
AT1G30570
|
HERK2, hercules receptor kinase 2 |
-0.413 |
-0.065 |
| 470 |
246885_at |
AT5G26230
|
MAKR1, membrane-associated kinase regulator 1 |
-0.411 |
-0.091 |
| 471 |
245864_at |
AT1G58070
|
unknown |
-0.409 |
-0.001 |
| 472 |
249037_at |
AT5G44130
|
FLA13, FASCICLIN-like arabinogalactan protein 13 precursor |
-0.407 |
-0.004 |
| 473 |
263942_at |
AT2G35860
|
FLA16, FASCICLIN-like arabinogalactan protein 16 precursor |
-0.405 |
-0.041 |
| 474 |
248427_at |
AT5G51750
|
ATSBT1.3, subtilase 1.3, SBT1.3, subtilase 1.3 |
-0.405 |
-0.002 |
| 475 |
267076_at |
AT2G41090
|
[Calcium-binding EF-hand family protein] |
-0.404 |
0.020 |
| 476 |
251423_at |
AT3G60550
|
CYCP3;2, cyclin p3;2 |
-0.404 |
0.061 |
| 477 |
257113_at |
AT3G20130
|
CYP705A22, cytochrome P450, family 705, subfamily A, polypeptide 22, GPS1, gravity persistence signal 1 |
-0.403 |
-0.110 |
| 478 |
264078_at |
AT2G28470
|
BGAL8, beta-galactosidase 8 |
-0.403 |
-0.098 |
| 479 |
247648_at |
AT5G60020
|
ATLAC17, LAC17, laccase 17 |
-0.403 |
-0.044 |
| 480 |
247731_at |
AT5G59590
|
UGT76E2, UDP-glucosyl transferase 76E2 |
-0.403 |
-0.007 |
| 481 |
256099_at |
AT1G13710
|
CYP78A5, cytochrome P450, family 78, subfamily A, polypeptide 5, KLU, KLUH |
-0.401 |
-0.048 |
| 482 |
254785_at |
AT4G12730
|
FLA2, FASCICLIN-like arabinogalactan 2 |
-0.397 |
-0.099 |
| 483 |
252896_at |
AT4G39480
|
CYP96A9, cytochrome P450, family 96, subfamily A, polypeptide 9 |
-0.397 |
-0.032 |
| 484 |
249986_at |
AT5G18460
|
unknown |
-0.397 |
0.006 |
| 485 |
260676_at |
AT1G19450
|
[Major facilitator superfamily protein] |
-0.395 |
-0.046 |
| 486 |
246144_at |
AT5G20110
|
[Dynein light chain type 1 family protein] |
-0.394 |
-0.061 |
| 487 |
263656_at |
AT1G04240
|
IAA3, indole-3-acetic acid inducible 3, SHY2, SHORT HYPOCOTYL 2 |
-0.394 |
-0.166 |
| 488 |
265066_at |
AT1G03870
|
FLA9, FASCICLIN-like arabinoogalactan 9 |
-0.392 |
-0.100 |