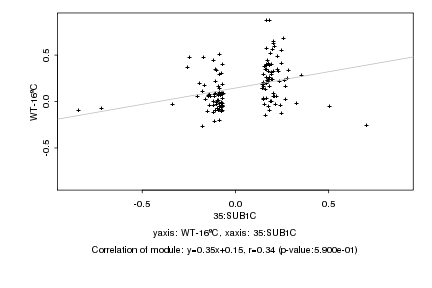
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
35:SUB1C |
Log2 signal ratio
WT-16ºC |
| 1 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.702 |
-0.255 |
| 2 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
0.504 |
-0.050 |
| 3 |
254056_at |
AT4G25250
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.353 |
0.282 |
| 4 |
249814_at |
AT5G23840
|
[MD-2-related lipid recognition domain-containing protein] |
0.327 |
-0.018 |
| 5 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.283 |
0.336 |
| 6 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
0.274 |
0.253 |
| 7 |
259964_at |
AT1G53680
|
ATGSTU28, glutathione S-transferase TAU 28, GSTU28, glutathione S-transferase TAU 28 |
0.266 |
0.168 |
| 8 |
266995_at |
AT2G34500
|
CYP710A1, cytochrome P450, family 710, subfamily A, polypeptide 1 |
0.266 |
0.023 |
| 9 |
256935_at |
AT3G22570
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.258 |
0.229 |
| 10 |
254909_at |
AT4G11210
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.256 |
0.688 |
| 11 |
249384_at |
AT5G39890
|
unknown |
0.246 |
-0.122 |
| 12 |
248727_at |
AT5G47990
|
CYP705A5, cytochrome P450, family 705, subfamily A, polypeptide 5, THAD, THALIAN-DIOL DESATURASE, THAD1, THALIAN-DIOL DESATURASE |
0.244 |
0.558 |
| 13 |
266669_at |
AT2G29750
|
UGT71C1, UDP-glucosyl transferase 71C1 |
0.242 |
0.416 |
| 14 |
247469_at |
AT5G62165
|
AGL42, AGAMOUS-like 42, FYF, FOREVER YOUNG FLOWER |
0.240 |
-0.034 |
| 15 |
255814_at |
AT1G19900
|
[glyoxal oxidase-related protein] |
0.236 |
0.221 |
| 16 |
254313_at |
AT4G22460
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.230 |
0.328 |
| 17 |
267307_at |
AT2G30210
|
LAC3, laccase 3 |
0.224 |
0.494 |
| 18 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
0.221 |
0.346 |
| 19 |
249205_at |
AT5G42600
|
MRN1, marneral synthase 1 |
0.218 |
0.055 |
| 20 |
245140_at |
AT2G45420
|
LBD18, LOB domain-containing protein 18 |
0.213 |
-0.023 |
| 21 |
249202_at |
AT5G42580
|
CYP705A12, cytochrome P450, family 705, subfamily A, polypeptide 12 |
0.209 |
0.060 |
| 22 |
264404_at |
AT2G25160
|
CYP82F1, cytochrome P450, family 82, subfamily F, polypeptide 1 |
0.205 |
0.596 |
| 23 |
254326_at |
AT4G22610
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.204 |
0.093 |
| 24 |
265031_at |
AT1G61590
|
[Protein kinase superfamily protein] |
0.202 |
0.329 |
| 25 |
266649_at |
AT2G25810
|
TIP4;1, tonoplast intrinsic protein 4;1 |
0.202 |
0.647 |
| 26 |
248545_at |
AT5G50260
|
CEP1, cysteine endopeptidase 1 |
0.200 |
0.060 |
| 27 |
255695_at |
AT4G00080
|
UNE11, unfertilized embryo sac 11 |
0.200 |
0.632 |
| 28 |
260560_at |
AT2G43590
|
[Chitinase family protein] |
0.194 |
0.242 |
| 29 |
253515_at |
AT4G31320
|
SAUR37, SMALL AUXIN UPREGULATED RNA 37 |
0.194 |
0.566 |
| 30 |
261956_at |
AT1G64590
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.192 |
0.322 |
| 31 |
266978_at |
AT2G39430
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.192 |
0.402 |
| 32 |
266711_at |
AT2G46740
|
AtGulLO5, GulLO5, L -gulono-1,4-lactone ( L -GulL) oxidase 5 |
0.191 |
0.244 |
| 33 |
262198_at |
AT1G53830
|
ATPME2, pectin methylesterase 2, PME2, pectin methylesterase 2 |
0.189 |
0.295 |
| 34 |
255150_at |
AT4G08160
|
[glycosyl hydrolase family 10 protein / carbohydrate-binding domain-containing protein] |
0.189 |
0.011 |
| 35 |
253454_at |
AT4G31875
|
unknown |
0.183 |
0.001 |
| 36 |
248252_at |
AT5G53250
|
AGP22, arabinogalactan protein 22, ATAGP22, ARABINOGALACTAN PROTEIN 22 |
0.183 |
0.527 |
| 37 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
0.182 |
-0.088 |
| 38 |
259388_at |
AT1G13420
|
ATST4B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 4B, ST4B, sulfotransferase 4B |
0.182 |
0.883 |
| 39 |
247586_at |
AT5G60660
|
PIP2;4, plasma membrane intrinsic protein 2;4, PIP2F |
0.180 |
0.264 |
| 40 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
0.179 |
0.393 |
| 41 |
253527_at |
AT4G31470
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.178 |
0.166 |
| 42 |
267094_at |
AT2G38080
|
ATLMCO4, ARABIDOPSIS LACCASE-LIKE MULTICOPPER OXIDASE 4, IRX12, IRREGULAR XYLEM 12, LAC4, LACCASE 4, LMCO4, LACCASE-LIKE MULTICOPPER OXIDASE 4 |
0.175 |
-0.044 |
| 43 |
259443_at |
AT1G02360
|
[Chitinase family protein] |
0.174 |
0.333 |
| 44 |
254828_at |
AT4G12550
|
AIR1, Auxin-Induced in Root cultures 1 |
0.174 |
0.216 |
| 45 |
263437_at |
AT2G28670
|
ESB1, ENHANCED SUBERIN 1 |
0.174 |
0.417 |
| 46 |
266920_at |
AT2G45750
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.174 |
0.245 |
| 47 |
249767_at |
AT5G24090
|
ATCHIA, chitinase A, CHIA, chitinase A |
0.171 |
0.228 |
| 48 |
257774_at |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
0.170 |
0.448 |
| 49 |
265355_at |
AT2G16760
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.167 |
0.236 |
| 50 |
246197_at |
AT4G37010
|
CEN2, centrin 2 |
0.166 |
0.392 |
| 51 |
261999_at |
AT1G33800
|
AtGXMT1, GXM1, glucuronoxylan methyltransferase 1, GXMT1, glucuronoxylan methyltransferase 1 |
0.164 |
0.408 |
| 52 |
266356_at |
AT2G32300
|
UCC1, uclacyanin 1 |
0.163 |
0.571 |
| 53 |
253667_at |
AT4G30170
|
[Peroxidase family protein] |
0.163 |
0.879 |
| 54 |
264716_at |
AT1G70170
|
MMP, matrix metalloproteinase |
0.162 |
0.269 |
| 55 |
246993_at |
AT5G67450
|
AZF1, zinc-finger protein 1, ZF1, zinc-finger protein 1 |
0.162 |
0.201 |
| 56 |
249812_at |
AT5G23830
|
[MD-2-related lipid recognition domain-containing protein] |
0.162 |
0.352 |
| 57 |
251012_at |
AT5G02580
|
unknown |
0.162 |
0.042 |
| 58 |
247467_at |
AT5G62130
|
[Per1-like family protein] |
0.160 |
-0.146 |
| 59 |
252209_at |
AT3G50400
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.158 |
0.347 |
| 60 |
245550_at |
AT4G15330
|
CYP705A1, cytochrome P450, family 705, subfamily A, polypeptide 1 |
0.156 |
0.138 |
| 61 |
258905_at |
AT3G06390
|
[Uncharacterised protein family (UPF0497)] |
0.154 |
0.380 |
| 62 |
264680_at |
AT1G65510
|
unknown |
0.151 |
-0.028 |
| 63 |
251723_at |
AT3G56230
|
[BTB/POZ domain-containing protein] |
0.150 |
0.194 |
| 64 |
259120_at |
AT3G02240
|
GLV4, GOLVEN 4, RGF7, root meristem growth factor 7 |
0.147 |
0.164 |
| 65 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
0.147 |
0.295 |
| 66 |
249031_at |
AT5G44900
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.146 |
0.028 |
| 67 |
261509_at |
AT1G71740
|
unknown |
0.146 |
0.208 |
| 68 |
262887_at |
AT1G14780
|
[MAC/Perforin domain-containing protein] |
0.146 |
0.037 |
| 69 |
262616_at |
AT1G06620
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.145 |
0.142 |
| 70 |
256900_at |
AT3G24670
|
[Pectin lyase-like superfamily protein] |
0.145 |
0.143 |
| 71 |
246373_at |
AT1G51860
|
[Leucine-rich repeat protein kinase family protein] |
0.144 |
0.192 |
| 72 |
266458_at |
AT2G47710
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.846 |
-0.087 |
| 73 |
266513_at |
AT2G47700
|
RFI2, RED AND FAR-RED INSENSITIVE 2 |
-0.719 |
-0.065 |
| 74 |
266461_at |
AT2G47730
|
ATGSTF5, GLUTATHIONE S-TRANSFERASE (CLASS PHI) 5, ATGSTF8, Arabidopsis thaliana glutathione S-transferase phi 8, GST6, GSTF8, glutathione S-transferase phi 8 |
-0.342 |
-0.024 |
| 75 |
254964_at |
AT4G11080
|
3xHMG-box1, 3xHigh Mobility Group-box1 |
-0.258 |
0.368 |
| 76 |
254550_at |
AT4G19690
|
ATIRT1, ARABIDOPSIS IRON-REGULATED TRANSPORTER 1, IRT1, iron-regulated transporter 1 |
-0.250 |
0.480 |
| 77 |
252938_at |
AT4G39190
|
unknown |
-0.204 |
0.060 |
| 78 |
247878_at |
AT5G57760
|
unknown |
-0.194 |
0.196 |
| 79 |
258935_at |
AT3G10120
|
unknown |
-0.182 |
-0.268 |
| 80 |
254663_at |
AT4G18290
|
KAT2, potassium channel in Arabidopsis thaliana 2 |
-0.178 |
0.113 |
| 81 |
262081_at |
AT1G59540
|
ZCF125 |
-0.174 |
0.484 |
| 82 |
250043_at |
AT5G18430
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.166 |
0.178 |
| 83 |
248070_at |
AT5G55660
|
[DEK domain-containing chromatin associated protein] |
-0.164 |
0.026 |
| 84 |
249271_at |
AT5G41790
|
CIP1, COP1-interactive protein 1 |
-0.151 |
-0.098 |
| 85 |
248623_at |
AT5G49170
|
unknown |
-0.149 |
0.068 |
| 86 |
256741_at |
AT3G29375
|
[XH domain-containing protein] |
-0.149 |
0.063 |
| 87 |
247331_at |
AT5G63530
|
ATFP3, ARABIDOPSIS THALIANA FARNESYLATED PROTEIN 3, FP3, farnesylated protein 3 |
-0.144 |
0.081 |
| 88 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
-0.140 |
-0.035 |
| 89 |
254675_at |
AT4G18465
|
[RNA helicase family protein] |
-0.139 |
0.064 |
| 90 |
255403_at |
AT4G03400
|
DFL2, DWARF IN LIGHT 2, GH3-10 |
-0.123 |
-0.033 |
| 91 |
246557_at |
AT5G15510
|
[TPX2 (targeting protein for Xklp2) protein family] |
-0.122 |
0.443 |
| 92 |
248539_at |
AT5G50130
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.121 |
-0.117 |
| 93 |
249843_at |
AT5G23570
|
ATSGS3, SUPPRESSOR OF GENE SILENCING 3, SGS3, SUPPRESSOR OF GENE SILENCING 3 |
-0.118 |
0.010 |
| 94 |
262040_at |
AT1G80080
|
AtRLP17, Receptor Like Protein 17, TMM, TOO MANY MOUTHS |
-0.116 |
0.083 |
| 95 |
261447_at |
AT1G21160
|
[eukaryotic translation initiation factor 2 (eIF-2) family protein] |
-0.113 |
-0.208 |
| 96 |
266508_at |
AT2G47920
|
NET3C, Networked 3C |
-0.113 |
0.064 |
| 97 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
-0.112 |
-0.093 |
| 98 |
253514_at |
AT4G31805
|
POLAR, POLAR LOCALIZATION DURING ASYMMETRIC DIVISION AND REDISTRIBUTION |
-0.111 |
0.216 |
| 99 |
258117_at |
AT3G14700
|
[SART-1 family] |
-0.109 |
-0.088 |
| 100 |
245461_at |
AT4G17000
|
unknown |
-0.108 |
0.350 |
| 101 |
262538_at |
AT1G17140
|
ICR1, interactor of constitutive active rops 1, RIP1, ROP INTERACTIVE PARTNER 1 |
-0.108 |
0.097 |
| 102 |
252089_at |
AT3G52110
|
unknown |
-0.105 |
0.344 |
| 103 |
262773_at |
AT1G13220
|
CRWN2, CROWDED NUCLEI 2, LINC2, LITTLE NUCLEI2 |
-0.104 |
-0.007 |
| 104 |
260876_at |
AT1G21460
|
AtSWEET1, SWEET1 |
-0.102 |
-0.026 |
| 105 |
257646_at |
AT3G25740
|
MAP1B, methionine aminopeptidase 1C, MAP1C, METHIONINE AMINOPEPTIDASE 1C |
-0.101 |
-0.072 |
| 106 |
247340_at |
AT5G63700
|
[zinc ion binding] |
-0.097 |
0.018 |
| 107 |
261325_at |
AT1G44780
|
unknown |
-0.095 |
0.082 |
| 108 |
259142_at |
AT3G10200
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.092 |
0.019 |
| 109 |
248710_at |
AT5G48480
|
[Lactoylglutathione lyase / glyoxalase I family protein] |
-0.092 |
0.068 |
| 110 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
-0.092 |
-0.086 |
| 111 |
253788_at |
AT4G28680
|
AtTYDC, TYDC, L-tyrosine decarboxylase, TYRDC, L-tyrosine decarboxylase, TYRDC1, L-TYROSINE DECARBOXYLASE 1 |
-0.092 |
0.168 |
| 112 |
257389_at |
AT2G17970
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.089 |
-0.078 |
| 113 |
247603_at |
AT5G60930
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.089 |
0.292 |
| 114 |
249184_at |
AT5G43020
|
[Leucine-rich repeat protein kinase family protein] |
-0.088 |
0.104 |
| 115 |
258098_at |
AT3G23670
|
KINESIN-12B, PAKRP1L |
-0.088 |
0.511 |
| 116 |
248807_at |
AT5G47500
|
PME5, pectin methylesterase 5 |
-0.087 |
0.150 |
| 117 |
264166_at |
AT1G65370
|
[TRAF-like family protein] |
-0.087 |
-0.197 |
| 118 |
266727_at |
AT2G03150
|
emb1579, embryo defective 1579 |
-0.086 |
-0.018 |
| 119 |
256111_at |
AT1G16820
|
[vacuolar ATP synthase catalytic subunit-related / V-ATPase-related / vacuolar proton pump-related] |
-0.086 |
0.080 |
| 120 |
253331_at |
AT4G33490
|
[Eukaryotic aspartyl protease family protein] |
-0.086 |
-0.048 |
| 121 |
258120_at |
AT3G14730
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.082 |
0.073 |
| 122 |
258376_at |
AT3G17680
|
[Kinase interacting (KIP1-like) family protein] |
-0.080 |
0.302 |
| 123 |
251946_at |
AT3G53540
|
TRM19, TON1 Recruiting Motif 19 |
-0.079 |
-0.002 |
| 124 |
260660_at |
AT1G19485
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.079 |
-0.101 |
| 125 |
267205_at |
AT2G30820
|
unknown |
-0.079 |
0.085 |
| 126 |
247812_at |
AT5G58390
|
[Peroxidase superfamily protein] |
-0.079 |
-0.039 |
| 127 |
256653_at |
AT3G18870
|
[Mitochondrial transcription termination factor family protein] |
-0.076 |
-0.105 |
| 128 |
251224_at |
AT3G62620
|
[sucrose-phosphatase-related] |
-0.076 |
-0.011 |
| 129 |
252911_at |
AT4G39510
|
CYP96A12, cytochrome P450, family 96, subfamily A, polypeptide 12 |
-0.076 |
0.095 |
| 130 |
250255_at |
AT5G13730
|
SIG4, sigma factor 4, SIGD |
-0.075 |
-0.056 |
| 131 |
251826_at |
AT3G55110
|
ABCG18, ATP-binding cassette G18 |
-0.074 |
0.091 |
| 132 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.074 |
0.184 |
| 133 |
260801_at |
AT1G78430
|
RIP4, ROP interactive partner 4 |
-0.072 |
0.402 |
| 134 |
262281_at |
AT1G68570
|
AtNPF3.1, NPF3.1, NRT1/ PTR family 3.1 |
-0.072 |
-0.011 |
| 135 |
251458_at |
AT3G60170
|
[copia-like retrotransposon family, has a 7.6e-229 P-value blast match to gb|AAO73527.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
-0.071 |
-0.033 |
| 136 |
250187_at |
AT5G14370
|
[CCT motif family protein] |
-0.071 |
-0.050 |
| 137 |
245417_at |
AT4G17360
|
[Formyl transferase] |
-0.071 |
-0.093 |
| 138 |
250245_at |
AT5G13690
|
CYL1, CYCLOPS 1, NAGLU, N-ACETYL-GLUCOSAMINIDASE |
-0.071 |
-0.050 |
| 139 |
256452_at |
AT1G75240
|
AtHB33, homeobox protein 33, HB33, homeobox protein 33, ZHD5, zinc-finger homeodomain 5 |
-0.070 |
0.040 |
| 140 |
251299_at |
AT3G61950
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.070 |
0.076 |
| 141 |
266512_at |
AT2G47690
|
[NADH-ubiquinone oxidoreductase-related] |
-0.069 |
0.097 |