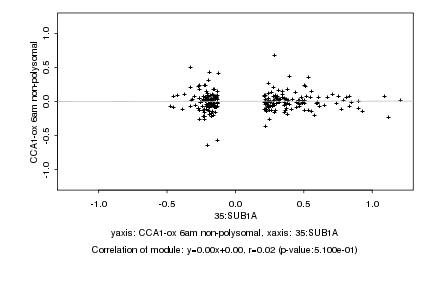
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
35:SUB1A |
Log2 signal ratio
CCA1-ox 6am non-polysomal |
| 1 |
266393_at |
AT2G41260
|
ATM17, M17 |
1.204 |
0.017 |
| 2 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
1.113 |
-0.234 |
| 3 |
249082_at |
AT5G44120
|
ATCRA1, CRUCIFERINA, CRA1, CRUCIFERINA, CRU1 |
1.087 |
0.079 |
| 4 |
249353_at |
AT5G40420
|
OLE2, OLEOSIN 2, OLEO2, oleosin 2 |
0.927 |
-0.140 |
| 5 |
256814_at |
AT3G21370
|
BGLU19, beta glucosidase 19 |
0.897 |
-0.098 |
| 6 |
260716_at |
AT1G48130
|
ATPER1, 1-cysteine peroxiredoxin 1, PER1, 1-cysteine peroxiredoxin 1 |
0.895 |
0.004 |
| 7 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
0.843 |
-0.001 |
| 8 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
0.828 |
0.084 |
| 9 |
266544_at |
AT2G35300
|
AtLEA4-2, late embryogenesis abundant 4-2, LEA18, late embryogenesis abundant 18, LEA4-2, late embryogenesis abundant 4-2 |
0.827 |
-0.061 |
| 10 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.811 |
0.073 |
| 11 |
258224_at |
AT3G15670
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.787 |
0.021 |
| 12 |
258347_at |
AT3G17520
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.773 |
-0.117 |
| 13 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
0.751 |
0.084 |
| 14 |
261339_at |
AT1G35710
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.732 |
-0.015 |
| 15 |
264079_at |
AT2G28490
|
[RmlC-like cupins superfamily protein] |
0.706 |
0.113 |
| 16 |
252549_at |
AT3G45860
|
CRK4, cysteine-rich RLK (RECEPTOR-like protein kinase) 4 |
0.673 |
0.062 |
| 17 |
247437_at |
AT5G62490
|
ATHVA22B, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE B, HVA22B, HVA22 homologue B |
0.648 |
-0.050 |
| 18 |
260088_at |
AT1G73190
|
ALPHA-TIP, ALPHA-TONOPLAST INTRINSIC PROTEIN, TIP3;1 |
0.610 |
-0.069 |
| 19 |
266654_at |
AT2G25890
|
[Oleosin family protein] |
0.602 |
0.073 |
| 20 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
0.596 |
-0.018 |
| 21 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
0.595 |
-0.002 |
| 22 |
259615_at |
AT1G47980
|
unknown |
0.589 |
-0.015 |
| 23 |
259579_at |
AT1G28010
|
ABCB14, ATP-binding cassette B14, ATABCB14, Arabidopsis thaliana ATP-binding cassette B14, MDR12, multi-drug resistance 12, PGP14, P-glycoprotein 14 |
0.571 |
-0.200 |
| 24 |
257184_at |
AT3G13090
|
ABCC6, ATP-binding cassette C6, ATMRP8, multidrug resistance-associated protein 8, MRP8, multidrug resistance-associated protein 8 |
0.556 |
0.154 |
| 25 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
0.550 |
-0.140 |
| 26 |
256464_at |
AT1G32560
|
AtLEA4-1, Late Embryogenesis Abundant 4-1, LEA4-1, Late Embryogenesis Abundant 4-1 |
0.547 |
0.066 |
| 27 |
252437_at |
AT3G47380
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.529 |
-0.130 |
| 28 |
251838_at |
AT3G54940
|
[Papain family cysteine protease] |
0.529 |
0.361 |
| 29 |
256053_at |
AT1G07260
|
UGT71C3, UDP-glucosyl transferase 71C3 |
0.513 |
0.086 |
| 30 |
250493_at |
AT5G09800
|
[ARM repeat superfamily protein] |
0.509 |
0.221 |
| 31 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.504 |
0.023 |
| 32 |
254271_at |
AT4G23150
|
CRK7, cysteine-rich RLK (RECEPTOR-like protein kinase) 7 |
0.503 |
0.246 |
| 33 |
265672_at |
AT2G31980
|
AtCYS2, PHYTOCYSTATIN 2, CYS2, PHYTOCYSTATIN 2 |
0.499 |
-0.025 |
| 34 |
250624_at |
AT5G07330
|
unknown |
0.498 |
-0.125 |
| 35 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
0.491 |
-0.003 |
| 36 |
263852_at |
AT2G04450
|
ATNUDT6, nudix hydrolase homolog 6, ATNUDX6, Arabidopsis thaliana nucleoside diphosphate linked to some moiety X 6, NUDT6, nudix hydrolase homolog 6, NUDX6, nucleoside diphosphates linked to some moiety X 6 |
0.487 |
0.029 |
| 37 |
267546_at |
AT2G32680
|
AtRLP23, receptor like protein 23, RLP23, receptor like protein 23 |
0.487 |
-0.019 |
| 38 |
264612_at |
AT1G04560
|
[AWPM-19-like family protein] |
0.483 |
-0.014 |
| 39 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
0.476 |
0.069 |
| 40 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
0.464 |
0.018 |
| 41 |
266143_at |
AT2G38905
|
[Low temperature and salt responsive protein family] |
0.463 |
-0.065 |
| 42 |
266462_at |
AT2G47770
|
ATTSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related, TSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related |
0.463 |
-0.032 |
| 43 |
254095_at |
AT4G25140
|
OLE1, OLEOSIN 1, OLEO1, oleosin 1 |
0.461 |
0.023 |
| 44 |
250868_at |
AT5G03860
|
MLS, malate synthase |
0.457 |
-0.018 |
| 45 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
0.446 |
-0.005 |
| 46 |
248169_at |
AT5G54610
|
ANK, ankyrin, BDA1, bian da 2 (becoming big in Chinese) |
0.446 |
-0.083 |
| 47 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
0.435 |
0.146 |
| 48 |
260904_at |
AT1G02450
|
NIMIN-1, NIMIN1, NIM1-interacting 1 |
0.414 |
0.032 |
| 49 |
248777_at |
AT5G47920
|
unknown |
0.406 |
-0.113 |
| 50 |
254293_at |
AT4G23060
|
IQD22, IQ-domain 22 |
0.393 |
0.370 |
| 51 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
0.389 |
-0.038 |
| 52 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
0.385 |
0.021 |
| 53 |
267101_at |
AT2G41480
|
AtPRX25, PRX25, peroxidase 25 |
0.380 |
0.187 |
| 54 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
0.379 |
-0.178 |
| 55 |
252060_at |
AT3G52430
|
ATPAD4, ARABIDOPSIS PHYTOALEXIN DEFICIENT 4, PAD4, PHYTOALEXIN DEFICIENT 4 |
0.376 |
0.023 |
| 56 |
254255_at |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
0.374 |
-0.027 |
| 57 |
260917_at |
AT1G02700
|
unknown |
0.371 |
-0.099 |
| 58 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
0.369 |
0.033 |
| 59 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
0.365 |
0.032 |
| 60 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
0.364 |
-0.123 |
| 61 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.360 |
-0.043 |
| 62 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.358 |
0.054 |
| 63 |
262585_at |
AT1G15460
|
ATBOR4, ARABIDOPSIS THALIANA REQUIRES HIGH BORON 4, BOR4, REQUIRES HIGH BORON 4 |
0.358 |
-0.153 |
| 64 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.358 |
-0.004 |
| 65 |
265856_at |
AT2G42430
|
ASL18, ASYMMETRIC LEAVES2-LIKE 18, LBD16, lateral organ boundaries-domain 16 |
0.357 |
-0.052 |
| 66 |
259616_at |
AT1G47960
|
ATC/VIF1, CELL WALL / VACUOLAR INHIBITOR OF FRUCTOSIDASE 1, C/VIF1, cell wall / vacuolar inhibitor of fructosidase 1 |
0.355 |
-0.044 |
| 67 |
266995_at |
AT2G34500
|
CYP710A1, cytochrome P450, family 710, subfamily A, polypeptide 1 |
0.344 |
0.155 |
| 68 |
259167_at |
AT3G01570
|
[Oleosin family protein] |
0.344 |
0.070 |
| 69 |
267384_at |
AT2G44370
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.342 |
0.113 |
| 70 |
255341_at |
AT4G04500
|
CRK37, cysteine-rich RLK (RECEPTOR-like protein kinase) 37 |
0.323 |
0.031 |
| 71 |
257130_at |
AT3G20210
|
DELTA-VPE, delta vacuolar processing enzyme, DELTAVPE, DELTA VACUOLAR PROCESSING ENZYME |
0.316 |
0.044 |
| 72 |
263175_at |
AT1G05510
|
OBAP1a, Oil Body-Associated Protein 1a |
0.315 |
-0.008 |
| 73 |
245244_at |
AT1G44350
|
ILL6, IAA-leucine resistant (ILR)-like gene 6 |
0.309 |
-0.001 |
| 74 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
0.309 |
0.017 |
| 75 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
0.308 |
0.173 |
| 76 |
245335_at |
AT4G16160
|
ATOEP16-2, ATOEP16-S |
0.306 |
0.082 |
| 77 |
257100_at |
AT3G25010
|
AtRLP41, receptor like protein 41, RLP41, receptor like protein 41 |
0.305 |
0.041 |
| 78 |
250468_at |
AT5G10120
|
[Ethylene insensitive 3 family protein] |
0.299 |
-0.021 |
| 79 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
0.298 |
0.071 |
| 80 |
252921_at |
AT4G39030
|
EDS5, ENHANCED DISEASE SUSCEPTIBILITY 5, SCORD3, susceptible to coronatine-deficient Pst DC3000 3, SID1, SALICYLIC ACID INDUCTION DEFICIENT 1 |
0.294 |
0.053 |
| 81 |
263385_at |
AT2G40170
|
ATEM6, ARABIDOPSIS EARLY METHIONINE-LABELLED 6, EM6, EARLY METHIONINE-LABELLED 6, GEA6, LATE EMBRYOGENESIS ABUNDANT 6 |
0.292 |
-0.119 |
| 82 |
262838_at |
AT1G14960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.290 |
0.099 |
| 83 |
248178_at |
AT5G54370
|
[Late embryogenesis abundant (LEA) protein-related] |
0.284 |
-0.050 |
| 84 |
255348_at |
AT4G03820
|
unknown |
0.284 |
0.028 |
| 85 |
248352_at |
AT5G52300
|
LTI65, LOW-TEMPERATURE-INDUCED 65, RD29B, RESPONSIVE TO DESSICATION 29B |
0.283 |
0.690 |
| 86 |
250780_at |
AT5G05290
|
ATEXP2, ATEXPA2, expansin A2, ATHEXP ALPHA 1.12, EXP2, EXPANSIN 2, EXPA2, expansin A2 |
0.283 |
-0.148 |
| 87 |
251580_at |
AT3G58450
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.276 |
-0.016 |
| 88 |
251130_at |
AT5G01180
|
AtNPF8.2, ATPTR5, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 5, NPF8.2, NRT1/ PTR family 8.2, PTR5, peptide transporter 5 |
0.275 |
0.079 |
| 89 |
266965_at |
AT2G39510
|
UMAMIT14, Usually multiple acids move in and out Transporters 14 |
0.272 |
0.206 |
| 90 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
0.271 |
0.024 |
| 91 |
254056_at |
AT4G25250
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.269 |
-0.073 |
| 92 |
253880_at |
AT4G27590
|
[Heavy metal transport/detoxification superfamily protein ] |
0.265 |
-0.057 |
| 93 |
248564_at |
AT5G49700
|
AHL17, AT-hook motif nuclear localized protein 17 |
0.262 |
-0.126 |
| 94 |
252209_at |
AT3G50400
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.261 |
0.133 |
| 95 |
245794_at |
AT1G32170
|
XTH30, xyloglucan endotransglucosylase/hydrolase 30, XTR4, xyloglucan endotransglycosylase 4 |
0.256 |
-0.065 |
| 96 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
0.252 |
0.013 |
| 97 |
265597_at |
AT2G20142
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.250 |
-0.009 |
| 98 |
247602_at |
AT5G60900
|
RLK1, receptor-like protein kinase 1 |
0.247 |
-0.263 |
| 99 |
247061_at |
AT5G66780
|
unknown |
0.244 |
-0.128 |
| 100 |
253525_at |
AT4G31330
|
unknown |
0.240 |
-0.054 |
| 101 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
0.239 |
0.028 |
| 102 |
266711_at |
AT2G46740
|
AtGulLO5, GulLO5, L -gulono-1,4-lactone ( L -GulL) oxidase 5 |
0.237 |
-0.085 |
| 103 |
262971_at |
AT1G75640
|
[Leucine-rich receptor-like protein kinase family protein] |
0.237 |
0.122 |
| 104 |
261033_at |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
0.236 |
0.266 |
| 105 |
266291_at |
AT2G29320
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.236 |
0.055 |
| 106 |
266316_at |
AT2G27080
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.235 |
-0.083 |
| 107 |
266689_at |
AT2G19930
|
[RNA-dependent RNA polymerase family protein] |
0.234 |
-0.034 |
| 108 |
257066_at |
AT3G18280
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.231 |
-0.070 |
| 109 |
249384_at |
AT5G39890
|
unknown |
0.231 |
-0.007 |
| 110 |
249096_at |
AT5G43910
|
[pfkB-like carbohydrate kinase family protein] |
0.225 |
-0.117 |
| 111 |
249485_at |
AT5G39020
|
[Malectin/receptor-like protein kinase family protein] |
0.225 |
-0.139 |
| 112 |
261568_at |
AT1G01030
|
NGA3, NGATHA3 |
0.222 |
-0.019 |
| 113 |
249127_at |
AT5G43500
|
ARP9, actin-related protein 9, ATARP9, actin-related protein 9 |
0.219 |
-0.103 |
| 114 |
251012_at |
AT5G02580
|
unknown |
0.218 |
0.035 |
| 115 |
248392_at |
AT5G52050
|
[MATE efflux family protein] |
0.218 |
-0.036 |
| 116 |
265189_at |
AT1G23840
|
unknown |
0.217 |
-0.358 |
| 117 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
0.215 |
0.089 |
| 118 |
246858_at |
AT5G25930
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.215 |
0.003 |
| 119 |
259213_at |
AT3G09010
|
[Protein kinase superfamily protein] |
0.212 |
-0.009 |
| 120 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
0.211 |
0.080 |
| 121 |
249202_at |
AT5G42580
|
CYP705A12, cytochrome P450, family 705, subfamily A, polypeptide 12 |
0.210 |
-0.116 |
| 122 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
0.210 |
0.099 |
| 123 |
245267_at |
AT4G14060
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.476 |
-0.065 |
| 124 |
256527_at |
AT1G66100
|
[Plant thionin] |
-0.455 |
0.079 |
| 125 |
251977_at |
AT3G53250
|
SAUR57, SMALL AUXIN UPREGULATED RNA 57 |
-0.455 |
-0.074 |
| 126 |
247474_at |
AT5G62280
|
unknown |
-0.428 |
0.102 |
| 127 |
249645_at |
AT5G36910
|
THI2.2, thionin 2.2 |
-0.394 |
-0.112 |
| 128 |
259104_at |
AT3G02170
|
LNG2, LONGIFOLIA2, TRM1, TON1 Recruiting Motif 1 |
-0.375 |
0.117 |
| 129 |
245088_at |
AT2G39850
|
[Subtilisin-like serine endopeptidase family protein] |
-0.335 |
0.213 |
| 130 |
265917_at |
AT2G15080
|
AtRLP19, receptor like protein 19, RLP19, receptor like protein 19 |
-0.334 |
-0.070 |
| 131 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.329 |
0.510 |
| 132 |
263549_at |
AT2G21650
|
ATRL2, ARABIDOPSIS RAD-LIKE 2, MEE3, MATERNAL EFFECT EMBRYO ARREST 3, RSM1, RADIALIS-LIKE SANT/MYB 1 |
-0.328 |
0.028 |
| 133 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.318 |
0.044 |
| 134 |
250043_at |
AT5G18430
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.300 |
0.076 |
| 135 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.295 |
-0.045 |
| 136 |
259373_at |
AT1G69160
|
unknown |
-0.275 |
-0.093 |
| 137 |
249894_at |
AT5G22580
|
[Stress responsive A/B Barrel Domain] |
-0.272 |
-0.092 |
| 138 |
255028_at |
AT4G09890
|
unknown |
-0.271 |
0.222 |
| 139 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-0.270 |
-0.254 |
| 140 |
259794_at |
AT1G64330
|
[myosin heavy chain-related] |
-0.269 |
0.047 |
| 141 |
254663_at |
AT4G18290
|
KAT2, potassium channel in Arabidopsis thaliana 2 |
-0.268 |
0.054 |
| 142 |
261844_at |
AT1G15940
|
[Tudor/PWWP/MBT superfamily protein] |
-0.267 |
0.246 |
| 143 |
248539_at |
AT5G50130
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.266 |
0.182 |
| 144 |
254964_at |
AT4G11080
|
3xHMG-box1, 3xHigh Mobility Group-box1 |
-0.263 |
-0.131 |
| 145 |
249546_at |
AT5G38150
|
PMI15, plastid movement impaired 15 |
-0.260 |
-0.007 |
| 146 |
247377_at |
AT5G63180
|
[Pectin lyase-like superfamily protein] |
-0.258 |
-0.003 |
| 147 |
266320_at |
AT2G46640
|
TAC1, Tiller Angle Control 1 |
-0.246 |
0.008 |
| 148 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.246 |
-0.069 |
| 149 |
259120_at |
AT3G02240
|
GLV4, GOLVEN 4, RGF7, root meristem growth factor 7 |
-0.239 |
-0.125 |
| 150 |
252319_at |
AT3G48710
|
[DEK domain-containing chromatin associated protein] |
-0.237 |
0.080 |
| 151 |
251402_at |
AT3G60290
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.235 |
0.044 |
| 152 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.235 |
0.017 |
| 153 |
249378_at |
AT5G40450
|
unknown |
-0.234 |
-0.215 |
| 154 |
266009_at |
AT2G37420
|
[ATP binding microtubule motor family protein] |
-0.231 |
-0.211 |
| 155 |
261553_at |
AT1G63420
|
unknown |
-0.230 |
-0.152 |
| 156 |
265265_at |
AT2G42900
|
[Plant basic secretory protein (BSP) family protein] |
-0.230 |
0.236 |
| 157 |
248684_at |
AT5G48485
|
DIR1, DEFECTIVE IN INDUCED RESISTANCE 1 |
-0.229 |
-0.114 |
| 158 |
249860_at |
AT5G22860
|
[Serine carboxypeptidase S28 family protein] |
-0.228 |
-0.251 |
| 159 |
254609_at |
AT4G18970
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.227 |
-0.206 |
| 160 |
263433_at |
AT2G22240
|
ATIPS2, INOSITOL 3-PHOSPHATE SYNTHASE 2, ATMIPS2, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 2, MIPS2, myo-inositol-1-phosphate synthase 2 |
-0.226 |
0.067 |
| 161 |
264319_at |
AT1G04110
|
AtSDD1, SDD1, STOMATAL DENSITY AND DISTRIBUTION 1 |
-0.223 |
0.003 |
| 162 |
253977_at |
AT4G26630
|
[DEK domain-containing chromatin associated protein] |
-0.221 |
0.061 |
| 163 |
248784_at |
AT5G47380
|
unknown |
-0.221 |
0.034 |
| 164 |
261491_at |
AT1G14350
|
AtMYB124, myb domain protein 124, FLP, FOUR LIPS, MYB124 |
-0.220 |
0.244 |
| 165 |
252765_at |
AT3G42800
|
unknown |
-0.217 |
0.159 |
| 166 |
262040_at |
AT1G80080
|
AtRLP17, Receptor Like Protein 17, TMM, TOO MANY MOUTHS |
-0.216 |
0.049 |
| 167 |
252078_at |
AT3G51740
|
IMK2, inflorescence meristem receptor-like kinase 2 |
-0.216 |
-0.060 |
| 168 |
259398_at |
AT1G17700
|
PRA1.F1, prenylated RAB acceptor 1.F1 |
-0.215 |
-0.034 |
| 169 |
259760_at |
AT1G77580
|
unknown |
-0.215 |
-0.105 |
| 170 |
260031_at |
AT1G68790
|
CRWN3, CROWDED NUCLEI 3, LINC3, LITTLE NUCLEI3 |
-0.211 |
0.131 |
| 171 |
247085_at |
AT5G66310
|
[ATP binding microtubule motor family protein] |
-0.211 |
0.022 |
| 172 |
265111_at |
AT1G62510
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.210 |
-0.635 |
| 173 |
258339_at |
AT3G16120
|
[Dynein light chain type 1 family protein] |
-0.210 |
0.065 |
| 174 |
252215_at |
AT3G50240
|
KICP-02 |
-0.207 |
-0.068 |
| 175 |
267460_at |
AT2G33810
|
SPL3, squamosa promoter binding protein-like 3 |
-0.206 |
-0.046 |
| 176 |
246562_at |
AT5G15580
|
LNG1, LONGIFOLIA1, TRM2, TON1 Recruiting Motif 2 |
-0.206 |
0.095 |
| 177 |
252322_at |
AT3G48550
|
unknown |
-0.204 |
-0.053 |
| 178 |
247812_at |
AT5G58390
|
[Peroxidase superfamily protein] |
-0.204 |
0.028 |
| 179 |
256875_at |
AT3G26330
|
CYP71B37, cytochrome P450, family 71, subfamily B, polypeptide 37 |
-0.203 |
-0.015 |
| 180 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
-0.202 |
-0.057 |
| 181 |
259888_at |
AT1G76350
|
NLP5, NIN-like protein 5 |
-0.201 |
-0.131 |
| 182 |
259839_at |
AT1G52190
|
AtNPF1.2, NPF1.2, NRT1/ PTR family 1.2, NRT1.11, nitrate transporter 1.11 |
-0.201 |
0.035 |
| 183 |
251372_at |
AT3G60520
|
unknown |
-0.199 |
0.323 |
| 184 |
245616_at |
AT4G14480
|
[Protein kinase superfamily protein] |
-0.198 |
0.029 |
| 185 |
267612_at |
AT2G26690
|
AtNPF6.2, NPF6.2, NRT1/ PTR family 6.2 |
-0.197 |
0.074 |
| 186 |
247288_at |
AT5G64330
|
JK218, NPH3, NON-PHOTOTROPIC HYPOCOTYL 3, RPT3, ROOT PHOTOTROPISM 3 |
-0.197 |
-0.032 |
| 187 |
258890_at |
AT3G05690
|
ATHAP2B, HEME ACTIVATOR PROTEIN (YEAST) HOMOLOG 2B, HAP2B, HEME ACTIVATOR PROTEIN (YEAST) HOMOLOG 2B, NF-YA2, nuclear factor Y, subunit A2, UNE8, UNFERTILIZED EMBRYO SAC 8 |
-0.196 |
-0.143 |
| 188 |
247903_at |
AT5G57340
|
unknown |
-0.195 |
-0.086 |
| 189 |
248247_at |
AT5G53210
|
SPCH, SPEECHLESS |
-0.195 |
-0.045 |
| 190 |
260475_at |
AT1G11080
|
scpl31, serine carboxypeptidase-like 31 |
-0.193 |
-0.178 |
| 191 |
258736_at |
AT3G05900
|
[neurofilament protein-related] |
-0.190 |
0.437 |
| 192 |
255403_at |
AT4G03400
|
DFL2, DWARF IN LIGHT 2, GH3-10 |
-0.188 |
-0.010 |
| 193 |
260371_at |
AT1G69690
|
AtTCP15, TCP15, TEOSINTE BRANCHED1/CYCLOIDEA/PCF 15 |
-0.187 |
0.091 |
| 194 |
247880_at |
AT5G57780
|
P1R1, P1R1 |
-0.185 |
0.043 |
| 195 |
246911_at |
AT5G25810
|
tny, TINY |
-0.182 |
0.054 |
| 196 |
248961_at |
AT5G45650
|
[subtilase family protein] |
-0.181 |
-0.068 |
| 197 |
256599_at |
AT3G14760
|
unknown |
-0.181 |
-0.031 |
| 198 |
251368_at |
AT3G61380
|
TRM14, TON1 Recruiting Motif 14 |
-0.180 |
-0.027 |
| 199 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
-0.180 |
0.022 |
| 200 |
258468_at |
AT3G06070
|
unknown |
-0.179 |
-0.207 |
| 201 |
264021_at |
AT2G21200
|
SAUR7, SMALL AUXIN UPREGULATED RNA 7 |
-0.176 |
-0.166 |
| 202 |
258139_at |
AT3G24520
|
AT-HSFC1, HSFC1, heat shock transcription factor C1 |
-0.173 |
-0.140 |
| 203 |
262162_at |
AT1G78020
|
unknown |
-0.173 |
-0.058 |
| 204 |
246476_at |
AT5G16730
|
[LOCATED IN: chloroplast] |
-0.171 |
0.116 |
| 205 |
267089_at |
AT2G38300
|
[myb-like HTH transcriptional regulator family protein] |
-0.170 |
0.017 |
| 206 |
260639_at |
AT1G53180
|
unknown |
-0.168 |
0.019 |
| 207 |
264525_at |
AT1G10060
|
ATBCAT-1, branched-chain amino acid transaminase 1, BCAT-1, branched-chain amino acid transaminase 1, BCAT1, branched-chain amino acid transferase 1 |
-0.165 |
0.177 |
| 208 |
249886_at |
AT5G22320
|
[Leucine-rich repeat (LRR) family protein] |
-0.165 |
-0.072 |
| 209 |
263545_at |
AT2G21560
|
unknown |
-0.164 |
-0.029 |
| 210 |
246700_at |
AT5G28030
|
DES1, L-cysteine desulfhydrase 1 |
-0.164 |
0.039 |
| 211 |
257021_at |
AT3G19710
|
BCAT4, branched-chain aminotransferase4 |
-0.163 |
-0.198 |
| 212 |
256873_at |
AT3G26310
|
CYP71B35, cytochrome P450, family 71, subfamily B, polypeptide 35 |
-0.162 |
-0.019 |
| 213 |
259815_at |
AT1G49870
|
unknown |
-0.161 |
0.094 |
| 214 |
264343_at |
AT1G11850
|
unknown |
-0.160 |
-0.025 |
| 215 |
255652_at |
AT4G00950
|
MEE47, maternal effect embryo arrest 47 |
-0.160 |
0.185 |
| 216 |
252419_at |
AT3G47510
|
unknown |
-0.158 |
-0.054 |
| 217 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.156 |
-0.051 |
| 218 |
252691_at |
AT3G44050
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.155 |
0.058 |
| 219 |
263841_at |
AT2G36870
|
AtXTH32, XTH32, xyloglucan endotransglucosylase/hydrolase 32 |
-0.155 |
0.061 |
| 220 |
253788_at |
AT4G28680
|
AtTYDC, TYDC, L-tyrosine decarboxylase, TYRDC, L-tyrosine decarboxylase, TYRDC1, L-TYROSINE DECARBOXYLASE 1 |
-0.155 |
0.081 |
| 221 |
255128_at |
AT4G08310
|
unknown |
-0.155 |
0.047 |
| 222 |
258367_at |
AT3G14370
|
WAG2 |
-0.155 |
-0.083 |
| 223 |
258796_at |
AT3G04630
|
WDL1, WVD2-like 1 |
-0.153 |
-0.077 |
| 224 |
250475_at |
AT5G10180
|
AST68, ARABIDOPSIS SULFATE TRANSPORTER 68, SULTR2;1, sulfate transporter 2;1 |
-0.153 |
-0.071 |
| 225 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
-0.150 |
-0.158 |
| 226 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
-0.149 |
0.030 |
| 227 |
248558_at |
AT5G49990
|
[Xanthine/uracil permease family protein] |
-0.148 |
-0.087 |
| 228 |
260494_at |
AT2G41820
|
PXC3, PXY/TDR-correlated 3 |
-0.146 |
0.088 |
| 229 |
263656_at |
AT1G04240
|
IAA3, indole-3-acetic acid inducible 3, SHY2, SHORT HYPOCOTYL 2 |
-0.144 |
0.056 |
| 230 |
245029_at |
AT2G26580
|
YAB5, YABBY5 |
-0.144 |
0.081 |
| 231 |
250665_at |
AT5G06980
|
LNK4, night light-inducible and clock-regulated 4 |
-0.144 |
0.067 |
| 232 |
265085_at |
AT1G03780
|
AtTPX2, TPX2, targeting protein for XKLP2 |
-0.142 |
-0.074 |
| 233 |
258476_at |
AT3G02400
|
[SMAD/FHA domain-containing protein ] |
-0.140 |
0.058 |
| 234 |
250233_at |
AT5G13460
|
IQD11, IQ-domain 11 |
-0.139 |
0.064 |
| 235 |
256652_at |
AT3G18850
|
LPAT5, lysophosphatidyl acyltransferase 5 |
-0.138 |
-0.021 |
| 236 |
251374_at |
AT3G60390
|
HAT3, homeobox-leucine zipper protein 3 |
-0.138 |
-0.568 |
| 237 |
259163_at |
AT3G01490
|
[Protein kinase superfamily protein] |
-0.134 |
0.157 |
| 238 |
253064_at |
AT4G37730
|
AtbZIP7, basic leucine-zipper 7, bZIP7, basic leucine-zipper 7 |
-0.133 |
-0.003 |
| 239 |
245479_at |
AT4G16140
|
[proline-rich family protein] |
-0.133 |
-0.064 |
| 240 |
248923_at |
AT5G45940
|
AtNUDT11, Arabidopsis thaliana nudix hydrolase homolog 11, AtNUDX11, Arabidopsis thaliana nudix hydrolase homolog 11, NUDT11, nudix hydrolase homolog 11, NUDX11, nudix hydrolase homolog 11 |
-0.133 |
0.043 |
| 241 |
262847_at |
AT1G14840
|
ATMAP70-4, microtubule-associated proteins 70-4, MAP70-4, microtubule-associated proteins 70-4 |
-0.133 |
-0.015 |
| 242 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
-0.132 |
0.090 |
| 243 |
255923_at |
AT1G22180
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.131 |
0.423 |