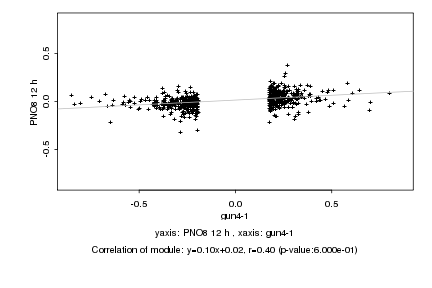
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
gun4-1 |
Log2 signal ratio
PNO8 12 h |
| 1 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.795 |
0.085 |
| 2 |
250182_at |
AT5G14470
|
AtGALK2, GALK2, galactokinase 2 |
0.700 |
-0.003 |
| 3 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
0.694 |
-0.093 |
| 4 |
264774_at |
AT1G22890
|
unknown |
0.641 |
0.117 |
| 5 |
264296_at |
AT1G78720
|
[SecY protein transport family protein] |
0.604 |
0.089 |
| 6 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
0.587 |
0.020 |
| 7 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
0.580 |
0.196 |
| 8 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
0.562 |
-0.044 |
| 9 |
245233_at |
AT4G25580
|
[CAP160 protein] |
0.508 |
-0.011 |
| 10 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
0.505 |
0.123 |
| 11 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
0.488 |
-0.044 |
| 12 |
260744_at |
AT1G15010
|
unknown |
0.479 |
0.122 |
| 13 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
0.476 |
0.098 |
| 14 |
260527_at |
AT2G47270
|
UPB1, UPBEAT1 |
0.465 |
0.022 |
| 15 |
246072_at |
AT5G20240
|
PI, PISTILLATA |
0.447 |
0.112 |
| 16 |
247135_at |
AT5G66260
|
SAUR11, SMALL AUXIN UPREGULATED RNA 11 |
0.441 |
0.021 |
| 17 |
258383_at |
AT3G15440
|
unknown |
0.435 |
0.012 |
| 18 |
258746_at |
AT3G05950
|
[RmlC-like cupins superfamily protein] |
0.434 |
0.013 |
| 19 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.431 |
0.046 |
| 20 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
0.417 |
0.034 |
| 21 |
261606_at |
AT1G49570
|
[Peroxidase superfamily protein] |
0.416 |
0.003 |
| 22 |
245970_at |
AT5G20710
|
BGAL7, beta-galactosidase 7 |
0.393 |
0.010 |
| 23 |
265644_at |
AT2G27380
|
ATEPR1, extensin proline-rich 1, EPR1, extensin proline-rich 1 |
0.387 |
0.062 |
| 24 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
0.386 |
0.158 |
| 25 |
264680_at |
AT1G65510
|
unknown |
0.386 |
0.077 |
| 26 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.384 |
0.078 |
| 27 |
259729_at |
AT1G77640
|
[Integrase-type DNA-binding superfamily protein] |
0.376 |
0.057 |
| 28 |
253628_at |
AT4G30280
|
ATXTH18, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18, XTH18, xyloglucan endotransglucosylase/hydrolase 18 |
0.376 |
-0.114 |
| 29 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.374 |
0.176 |
| 30 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
0.368 |
-0.030 |
| 31 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
0.355 |
0.032 |
| 32 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.354 |
0.115 |
| 33 |
248611_at |
AT5G49520
|
ATWRKY48, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 48, WRKY48, WRKY DNA-binding protein 48 |
0.344 |
0.062 |
| 34 |
252137_at |
AT3G50980
|
XERO1, dehydrin xero 1 |
0.343 |
0.059 |
| 35 |
265536_at |
AT2G15880
|
[Leucine-rich repeat (LRR) family protein] |
0.343 |
0.054 |
| 36 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.338 |
0.084 |
| 37 |
265732_at |
AT2G01300
|
unknown |
0.338 |
0.047 |
| 38 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
0.336 |
0.168 |
| 39 |
264124_at |
AT1G79360
|
ATOCT2, organic cation/carnitine transporter 2, OCT2, ORGANIC CATION TRANSPORTER 2, OCT2, organic cation/carnitine transporter 2 |
0.333 |
0.056 |
| 40 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.327 |
-0.114 |
| 41 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
0.326 |
-0.134 |
| 42 |
251226_at |
AT3G62680
|
ATPRP3, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 3, PRP3, proline-rich protein 3 |
0.325 |
-0.012 |
| 43 |
251647_at |
AT3G57770
|
[Protein kinase superfamily protein] |
0.324 |
0.033 |
| 44 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.324 |
0.107 |
| 45 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
0.323 |
0.132 |
| 46 |
263890_at |
AT2G37030
|
SAUR46, SMALL AUXIN UPREGULATED RNA 46 |
0.319 |
0.107 |
| 47 |
260623_at |
AT1G08090
|
ACH1, ATNRT2.1, NITRATE TRANSPORTER 2.1, ATNRT2:1, nitrate transporter 2:1, LIN1, LATERAL ROOT INITIATION 1, NRT2, NITRATE TRANSPORTER 2, NRT2.1, NITRATE TRANSPORTER 2.1, NRT2:1, nitrate transporter 2:1, NRT2;1AT |
0.319 |
0.064 |
| 48 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
0.319 |
0.076 |
| 49 |
264395_at |
AT1G12070
|
[Immunoglobulin E-set superfamily protein] |
0.318 |
0.086 |
| 50 |
251750_at |
AT3G55710
|
[UDP-Glycosyltransferase superfamily protein] |
0.318 |
0.030 |
| 51 |
256386_at |
AT1G66540
|
[Cytochrome P450 superfamily protein] |
0.317 |
-0.022 |
| 52 |
267083_at |
AT2G41100
|
ATCAL4, ARABIDOPSIS THALIANA CALMODULIN LIKE 4, TCH3, TOUCH 3 |
0.316 |
0.025 |
| 53 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.315 |
0.087 |
| 54 |
258923_at |
AT3G10450
|
SCPL7, serine carboxypeptidase-like 7 |
0.312 |
0.134 |
| 55 |
262977_at |
AT1G75490
|
[Integrase-type DNA-binding superfamily protein] |
0.310 |
0.037 |
| 56 |
254314_at |
AT4G22470
|
[protease inhibitor/seed storage/lipid transfer protein (LTP) family protein] |
0.310 |
-0.150 |
| 57 |
258873_at |
AT3G03240
|
[alpha/beta-Hydrolases superfamily protein] |
0.308 |
-0.017 |
| 58 |
256340_at |
AT1G72070
|
[Chaperone DnaJ-domain superfamily protein] |
0.307 |
0.057 |
| 59 |
264580_at |
AT1G05340
|
unknown |
0.306 |
0.158 |
| 60 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.305 |
0.082 |
| 61 |
264635_at |
AT1G65500
|
unknown |
0.302 |
-0.183 |
| 62 |
254197_at |
AT4G24040
|
ATTRE1, TRE1, trehalase 1 |
0.301 |
-0.001 |
| 63 |
258339_at |
AT3G16120
|
[Dynein light chain type 1 family protein] |
0.301 |
0.066 |
| 64 |
245543_at |
AT4G15260
|
[UDP-Glycosyltransferase superfamily protein] |
0.300 |
-0.018 |
| 65 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
0.299 |
-0.012 |
| 66 |
255285_at |
AT4G04630
|
unknown |
0.298 |
0.042 |
| 67 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
0.297 |
0.093 |
| 68 |
263281_at |
AT2G14160
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.296 |
0.039 |
| 69 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
0.295 |
-0.057 |
| 70 |
263593_at |
AT2G01860
|
EMB975, EMBRYO DEFECTIVE 975 |
0.294 |
-0.057 |
| 71 |
262549_at |
AT1G31290
|
AGO3, ARGONAUTE 3 |
0.290 |
0.085 |
| 72 |
250098_at |
AT5G17350
|
unknown |
0.290 |
0.019 |
| 73 |
247292_at |
AT5G64490
|
[ARM repeat superfamily protein] |
0.281 |
0.028 |
| 74 |
252833_at |
AT4G40090
|
AGP3, arabinogalactan protein 3 |
0.281 |
-0.019 |
| 75 |
261673_at |
AT1G18280
|
LTPG3, glycosylphosphatidylinositol-anchored lipid protein transfer 3 |
0.280 |
0.115 |
| 76 |
262640_at |
AT1G62760
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.280 |
0.026 |
| 77 |
244950_at |
ATMG00160
|
COX2, cytochrome oxidase 2 |
0.279 |
0.041 |
| 78 |
247848_at |
AT5G58120
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.276 |
0.119 |
| 79 |
261942_at |
AT1G22590
|
AGL87, AGAMOUS-like 87 |
0.274 |
-0.135 |
| 80 |
260656_at |
AT1G19380
|
unknown |
0.274 |
0.003 |
| 81 |
259566_at |
AT1G20520
|
unknown |
0.273 |
0.044 |
| 82 |
251974_at |
AT3G53200
|
AtMYB27, myb domain protein 27, MYB27, myb domain protein 27 |
0.273 |
0.033 |
| 83 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
0.272 |
-0.060 |
| 84 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
0.271 |
0.167 |
| 85 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
0.270 |
0.376 |
| 86 |
248347_at |
AT5G52250
|
EFO1, EARLY FLOWERING BY OVEREXPRESSION 1, RUP1, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 |
0.268 |
0.018 |
| 87 |
249091_at |
AT5G43860
|
ATCLH2, ARABIDOPSIS THALIANA CHLOROPHYLLASE 2, CLH2, chlorophyllase 2 |
0.268 |
0.009 |
| 88 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
0.267 |
0.105 |
| 89 |
258227_at |
AT3G15620
|
UVR3, UV REPAIR DEFECTIVE 3 |
0.267 |
0.034 |
| 90 |
252924_at |
AT4G39070
|
BBX20, B-box domain protein 20, BZS1, BZS1 |
0.266 |
0.019 |
| 91 |
261658_at |
AT1G50040
|
unknown |
0.263 |
0.107 |
| 92 |
258063_at |
AT3G14620
|
CYP72A8, cytochrome P450, family 72, subfamily A, polypeptide 8 |
0.260 |
0.091 |
| 93 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.259 |
0.046 |
| 94 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
0.258 |
0.150 |
| 95 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.257 |
0.006 |
| 96 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
0.257 |
0.302 |
| 97 |
267337_at |
AT2G39980
|
[HXXXD-type acyl-transferase family protein] |
0.257 |
-0.010 |
| 98 |
257690_at |
AT3G12830
|
SAUR72, SMALL AUXIN UPREGULATED 72 |
0.256 |
-0.014 |
| 99 |
253496_at |
AT4G31870
|
ATGPX7, GLUTATHIONE PEROXIDASE 7, GPX7, glutathione peroxidase 7 |
0.254 |
0.065 |
| 100 |
245349_at |
AT4G16690
|
ATMES16, ARABIDOPSIS THALIANA METHYL ESTERASE 16, MES16, methyl esterase 16 |
0.254 |
0.114 |
| 101 |
252539_at |
AT3G45730
|
unknown |
0.254 |
0.007 |
| 102 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
0.252 |
0.151 |
| 103 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
0.252 |
0.080 |
| 104 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.252 |
0.172 |
| 105 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.252 |
0.079 |
| 106 |
246018_at |
AT5G10695
|
unknown |
0.251 |
0.265 |
| 107 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
0.251 |
-0.025 |
| 108 |
257672_at |
AT3G20300
|
unknown |
0.250 |
-0.054 |
| 109 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.250 |
0.114 |
| 110 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.250 |
0.063 |
| 111 |
261934_at |
AT1G22400
|
ATUGT85A1, ARABIDOPSIS THALIANA UDP-GLUCOSYL TRANSFERASE 85A1, UGT85A1 |
0.248 |
0.037 |
| 112 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
0.247 |
0.103 |
| 113 |
252160_at |
AT3G50570
|
[hydroxyproline-rich glycoprotein family protein] |
0.247 |
-0.008 |
| 114 |
260774_at |
AT1G78290
|
SNRK2-8, SNF1-RELATED PROTEIN KINASE 2-8, SNRK2.8, SNF1-RELATED PROTEIN KINASE 2.8, SRK2C, SNF1-RELATED PROTEIN KINASE 2C |
0.247 |
0.124 |
| 115 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
0.246 |
0.090 |
| 116 |
265296_at |
AT2G14060
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.244 |
0.058 |
| 117 |
255521_at |
AT4G02280
|
ATSUS3, SUS3, sucrose synthase 3 |
0.243 |
0.077 |
| 118 |
251774_at |
AT3G55840
|
[Hs1pro-1 protein] |
0.243 |
-0.014 |
| 119 |
255064_at |
AT4G08950
|
EXO, EXORDIUM |
0.242 |
0.037 |
| 120 |
252462_at |
AT3G47250
|
unknown |
0.242 |
0.022 |
| 121 |
256970_at |
AT3G21090
|
ABCG15, ATP-binding cassette G15 |
0.241 |
0.029 |
| 122 |
256464_at |
AT1G32560
|
AtLEA4-1, Late Embryogenesis Abundant 4-1, LEA4-1, Late Embryogenesis Abundant 4-1 |
0.240 |
0.005 |
| 123 |
260205_at |
AT1G70700
|
JAZ9, JASMONATE-ZIM-DOMAIN PROTEIN 9, TIFY7 |
0.240 |
0.096 |
| 124 |
254770_at |
AT4G13340
|
LRX3, leucine-rich repeat/extensin 3 |
0.240 |
0.025 |
| 125 |
267451_at |
AT2G33710
|
[Integrase-type DNA-binding superfamily protein] |
0.239 |
0.076 |
| 126 |
255881_at |
AT1G67070
|
DIN9, DARK INDUCIBLE 9, PMI2, PHOSPHOMANNOSE ISOMERASE 2 |
0.238 |
0.063 |
| 127 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
0.238 |
-0.038 |
| 128 |
255733_at |
AT1G25400
|
unknown |
0.238 |
0.042 |
| 129 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.237 |
0.196 |
| 130 |
252136_at |
AT3G50770
|
CML41, calmodulin-like 41 |
0.237 |
0.050 |
| 131 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
0.235 |
-0.014 |
| 132 |
263106_at |
AT2G05160
|
[CCCH-type zinc fingerfamily protein with RNA-binding domain] |
0.235 |
0.051 |
| 133 |
264238_at |
AT1G54740
|
unknown |
0.233 |
0.086 |
| 134 |
250910_at |
AT5G03720
|
AT-HSFA3, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A3, HSFA3, heat shock transcription factor A3 |
0.232 |
0.112 |
| 135 |
245966_at |
AT5G19790
|
RAP2.11, related to AP2 11 |
0.232 |
-0.001 |
| 136 |
256420_at |
AT1G33475
|
[SNARE-like superfamily protein] |
0.231 |
0.021 |
| 137 |
245528_at |
AT4G15530
|
PPDK, pyruvate orthophosphate dikinase |
0.231 |
0.093 |
| 138 |
252387_at |
AT3G47800
|
[Galactose mutarotase-like superfamily protein] |
0.230 |
0.104 |
| 139 |
257666_at |
AT3G20270
|
[lipid-binding serum glycoprotein family protein] |
0.229 |
0.019 |
| 140 |
266265_at |
AT2G29340
|
[NAD-dependent epimerase/dehydratase family protein] |
0.228 |
0.007 |
| 141 |
258608_at |
AT3G03020
|
unknown |
0.228 |
-0.002 |
| 142 |
266278_at |
AT2G29300
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.226 |
0.040 |
| 143 |
265983_at |
AT2G18550
|
ATHB21, homeobox protein 21, HB-2, homeobox-2, HB21, homeobox protein 21 |
0.226 |
0.068 |
| 144 |
246943_at |
AT5G25440
|
[Protein kinase superfamily protein] |
0.226 |
-0.055 |
| 145 |
246464_at |
AT5G16980
|
[Zinc-binding dehydrogenase family protein] |
0.226 |
-0.066 |
| 146 |
259982_at |
AT1G76410
|
ATL8 |
0.226 |
0.112 |
| 147 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
0.225 |
-0.040 |
| 148 |
262705_at |
AT1G16260
|
[Wall-associated kinase family protein] |
0.225 |
0.094 |
| 149 |
266690_at |
AT2G19900
|
ATNADP-ME1, Arabidopsis thaliana NADP-malic enzyme 1, NADP-ME1, NADP-malic enzyme 1 |
0.223 |
0.069 |
| 150 |
250766_at |
AT5G05550
|
[sequence-specific DNA binding transcription factors] |
0.222 |
0.117 |
| 151 |
257401_at |
AT1G23550
|
SRO2, similar to RCD one 2 |
0.222 |
0.100 |
| 152 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
0.221 |
0.159 |
| 153 |
258049_at |
AT3G16220
|
[Predicted eukaryotic LigT] |
0.220 |
0.070 |
| 154 |
258983_at |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
0.220 |
0.020 |
| 155 |
247617_at |
AT5G60270
|
LecRK-I.7, L-type lectin receptor kinase I.7 |
0.219 |
0.075 |
| 156 |
264379_at |
AT2G25200
|
unknown |
0.219 |
0.171 |
| 157 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
0.219 |
-0.052 |
| 158 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
0.218 |
0.069 |
| 159 |
262412_at |
AT1G34760
|
GF14 OMICRON, GRF11, general regulatory factor 11, RHS5, ROOT HAIR SPECIFIC 5 |
0.218 |
0.010 |
| 160 |
262580_at |
AT1G15330
|
[Cystathionine beta-synthase (CBS) protein] |
0.218 |
0.006 |
| 161 |
267081_at |
AT2G41210
|
PIP5K5, phosphatidylinositol- 4-phosphate 5-kinase 5 |
0.217 |
0.031 |
| 162 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
0.217 |
0.069 |
| 163 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.216 |
-0.012 |
| 164 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
0.216 |
0.021 |
| 165 |
245737_at |
AT1G44160
|
[HSP40/DnaJ peptide-binding protein] |
0.214 |
0.074 |
| 166 |
265203_at |
AT2G36630
|
[Sulfite exporter TauE/SafE family protein] |
0.214 |
0.048 |
| 167 |
253281_at |
AT4G34138
|
UGT73B1, UDP-glucosyl transferase 73B1 |
0.213 |
0.094 |
| 168 |
259694_at |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
0.213 |
0.048 |
| 169 |
260664_at |
AT1G19510
|
ATRL5, RAD-like 5, RL5, RAD-like 5, RSM4, RADIALIS-LIKE SANT/MYB 4 |
0.213 |
-0.007 |
| 170 |
259911_at |
AT1G72680
|
ATCAD1, CINNAMYL ALCOHOL DEHYDROGENASE 1, CAD1, cinnamyl-alcohol dehydrogenase |
0.212 |
-0.004 |
| 171 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.212 |
0.147 |
| 172 |
251842_at |
AT3G54580
|
[Proline-rich extensin-like family protein] |
0.210 |
-0.043 |
| 173 |
252167_at |
AT3G50560
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.210 |
0.085 |
| 174 |
256324_at |
AT1G66760
|
[MATE efflux family protein] |
0.209 |
0.105 |
| 175 |
257670_at |
AT3G20340
|
unknown |
0.209 |
0.064 |
| 176 |
250292_at |
AT5G13220
|
JAS1, JASMONATE-ASSOCIATED 1, JAZ10, jasmonate-zim-domain protein 10, TIFY9, TIFY DOMAIN PROTEIN 9 |
0.209 |
0.125 |
| 177 |
258275_at |
AT3G15760
|
unknown |
0.208 |
0.155 |
| 178 |
256122_at |
AT1G18180
|
[FUNCTIONS IN: oxidoreductase activity, acting on the CH-CH group of donors] |
0.208 |
0.051 |
| 179 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
0.208 |
-0.152 |
| 180 |
262952_at |
AT1G75770
|
unknown |
0.208 |
-0.032 |
| 181 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.208 |
-0.015 |
| 182 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.207 |
-0.155 |
| 183 |
250472_at |
AT5G10210
|
unknown |
0.207 |
-0.077 |
| 184 |
266695_at |
AT2G19810
|
AtOZF1, AtTZF2, OZF1, Oxidation-related Zinc Finger 1, TZF2, tandem zinc finger 2 |
0.207 |
0.047 |
| 185 |
262456_at |
AT1G11260
|
ATSTP1, SUGAR TRANSPORTER 1, STP1, sugar transporter 1 |
0.207 |
0.145 |
| 186 |
254638_at |
AT4G18740
|
[Rho termination factor] |
0.206 |
0.121 |
| 187 |
263376_at |
AT2G20520
|
FLA6, FASCICLIN-like arabinogalactan 6 |
0.206 |
-0.034 |
| 188 |
259479_at |
AT1G19020
|
unknown |
0.205 |
-0.032 |
| 189 |
254424_at |
AT4G21510
|
AtFBS2, FBS2, F-box stress induced 2 |
0.205 |
0.042 |
| 190 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
0.202 |
0.102 |
| 191 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
0.202 |
-0.012 |
| 192 |
250223_at |
AT5G14070
|
ROXY2 |
0.202 |
0.086 |
| 193 |
250550_at |
AT5G07870
|
[HXXXD-type acyl-transferase family protein] |
0.201 |
0.092 |
| 194 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.201 |
0.192 |
| 195 |
256632_at |
AT3G28330
|
[F-box family protein-related] |
0.201 |
0.120 |
| 196 |
246260_at |
AT1G31820
|
PUT1, POLYAMINE UPTAKE TRANSPORTER 1 |
0.201 |
0.132 |
| 197 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
0.201 |
0.078 |
| 198 |
264843_at |
AT1G03400
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.201 |
0.102 |
| 199 |
261936_at |
AT1G22600
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.201 |
0.135 |
| 200 |
246963_at |
AT5G24820
|
[Eukaryotic aspartyl protease family protein] |
0.199 |
-0.008 |
| 201 |
259629_at |
AT1G56510
|
ADR2, ACTIVATED DISEASE RESISTANCE 2, WRR4, WHITE RUST RESISTANCE 4 |
0.199 |
0.044 |
| 202 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
0.198 |
0.046 |
| 203 |
247109_at |
AT5G65870
|
ATPSK5, phytosulfokine 5 precursor, PSK5, PSK5, phytosulfokine 5 precursor |
0.198 |
0.130 |
| 204 |
251494_at |
AT3G59350
|
[Protein kinase superfamily protein] |
0.198 |
0.004 |
| 205 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
0.198 |
-0.139 |
| 206 |
250064_at |
AT5G17890
|
CHS3, CHILLING SENSITIVE 3, DAR4, DA1-related protein 4 |
0.197 |
-0.057 |
| 207 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
0.197 |
0.003 |
| 208 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
0.197 |
0.064 |
| 209 |
267036_at |
AT2G38465
|
unknown |
0.196 |
0.081 |
| 210 |
256740_at |
AT3G29330
|
unknown |
0.196 |
0.057 |
| 211 |
247675_at |
AT5G59940
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.195 |
0.046 |
| 212 |
245392_at |
AT4G15680
|
[Thioredoxin superfamily protein] |
0.195 |
0.104 |
| 213 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.195 |
0.005 |
| 214 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
0.195 |
0.127 |
| 215 |
258262_at |
AT3G15770
|
unknown |
0.195 |
0.152 |
| 216 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
0.195 |
0.193 |
| 217 |
260918_at |
AT1G21510
|
unknown |
0.194 |
-0.028 |
| 218 |
259172_at |
AT3G03630
|
CS26, cysteine synthase 26 |
0.193 |
-0.083 |
| 219 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
0.193 |
-0.000 |
| 220 |
251971_at |
AT3G53160
|
UGT73C7, UDP-glucosyl transferase 73C7 |
0.193 |
0.072 |
| 221 |
256633_at |
AT3G28340
|
GATL10, galacturonosyltransferase-like 10, GolS8, galactinol synthase 8 |
0.192 |
0.001 |
| 222 |
245906_at |
AT5G11070
|
unknown |
0.192 |
0.033 |
| 223 |
259615_at |
AT1G47980
|
unknown |
0.192 |
0.046 |
| 224 |
256815_at |
AT3G21380
|
[Mannose-binding lectin superfamily protein] |
0.192 |
-0.025 |
| 225 |
267035_at |
AT2G38400
|
AGT3, alanine:glyoxylate aminotransferase 3 |
0.191 |
0.037 |
| 226 |
264005_at |
AT2G22470
|
AGP2, arabinogalactan protein 2, ATAGP2 |
0.191 |
-0.020 |
| 227 |
254139_at |
AT4G24600
|
unknown |
0.191 |
0.001 |
| 228 |
244901_at |
ATMG00640
|
ORF25 |
0.190 |
-0.004 |
| 229 |
266203_at |
AT2G02230
|
AtPP2-B1, phloem protein 2-B1, PP2-B1, phloem protein 2-B1 |
0.190 |
-0.001 |
| 230 |
259725_at |
AT1G61065
|
unknown |
0.190 |
0.054 |
| 231 |
257026_at |
AT3G19200
|
unknown |
0.190 |
-0.030 |
| 232 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
0.190 |
0.080 |
| 233 |
252095_at |
AT3G51000
|
[alpha/beta-Hydrolases superfamily protein] |
0.189 |
0.025 |
| 234 |
261423_at |
AT1G18750
|
AGL65, AGAMOUS-like 65 |
0.188 |
0.099 |
| 235 |
253884_at |
AT4G27670
|
HSP21, heat shock protein 21 |
0.188 |
0.162 |
| 236 |
245383_at |
AT4G17810
|
[C2H2 and C2HC zinc fingers superfamily protein] |
0.188 |
0.021 |
| 237 |
259244_at |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
0.188 |
0.049 |
| 238 |
267181_at |
AT2G37760
|
AKR4C8, Aldo-keto reductase family 4 member C8 |
0.188 |
0.125 |
| 239 |
247951_at |
AT5G57240
|
ORP4C, OSBP(oxysterol binding protein)-related protein 4C |
0.187 |
0.170 |
| 240 |
264752_at |
AT1G23010
|
LPR1, Low Phosphate Root1 |
0.187 |
0.065 |
| 241 |
257737_at |
AT3G27440
|
UKL5, uridine kinase-like 5 |
0.187 |
-0.033 |
| 242 |
250683_x_at |
AT5G06640
|
EXT10, extensin 10 |
0.187 |
-0.010 |
| 243 |
259685_at |
AT1G63090
|
AtPP2-A11, phloem protein 2-A11, PP2-A11, phloem protein 2-A11 |
0.187 |
-0.029 |
| 244 |
251039_at |
AT5G02020
|
SIS, Salt Induced Serine rich |
0.187 |
0.147 |
| 245 |
247013_at |
AT5G67480
|
ATBT4, BT4, BTB and TAZ domain protein 4 |
0.187 |
-0.065 |
| 246 |
264066_at |
AT2G27880
|
AGO5, ARGONAUTE 5, AtAGO5 |
0.186 |
0.071 |
| 247 |
246502_at |
AT5G16240
|
[Plant stearoyl-acyl-carrier-protein desaturase family protein] |
0.186 |
-0.011 |
| 248 |
257869_at |
AT3G25160
|
[ER lumen protein retaining receptor family protein] |
0.186 |
0.051 |
| 249 |
265906_at |
AT2G25565
|
unknown |
0.185 |
-0.036 |
| 250 |
246025_at |
AT5G21150
|
AGO9, ARGONAUTE 9 |
0.185 |
0.054 |
| 251 |
251642_at |
AT3G57520
|
AtSIP2, seed imbibition 2, RS2, raffinose synthase 2, SIP2, seed imbibition 2 |
0.185 |
0.065 |
| 252 |
246703_at |
AT5G28080
|
WNK9 |
0.185 |
0.031 |
| 253 |
259700_at |
AT1G68980
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.185 |
0.003 |
| 254 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
0.185 |
0.120 |
| 255 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
0.185 |
0.040 |
| 256 |
263377_at |
AT2G40220
|
ABI4, ABA INSENSITIVE 4, ATABI4, GIN6, GLUCOSE INSENSITIVE 6, ISI3, IMPAIRED SUCROSE INDUCTION 3, SAN5, SALOBRENO 5, SIS5, SUGAR-INSENSITIVE 5, SUN6, SUCROSE UNCOUPLED 6 |
0.184 |
0.059 |
| 257 |
255140_x_at |
AT4G08410
|
[Proline-rich extensin-like family protein] |
0.184 |
0.010 |
| 258 |
266269_at |
AT2G29480
|
ATGSTU2, glutathione S-transferase tau 2, GST20, GLUTATHIONE S-TRANSFERASE 20, GSTU2, glutathione S-transferase tau 2 |
0.184 |
0.023 |
| 259 |
247524_at |
AT5G61440
|
ACHT5, atypical CYS HIS rich thioredoxin 5 |
0.183 |
0.116 |
| 260 |
247793_at |
AT5G58650
|
PSY1, plant peptide containing sulfated tyrosine 1 |
0.183 |
0.112 |
| 261 |
255381_at |
AT4G03510
|
ATRMA1, RMA1, RING membrane-anchor 1 |
0.183 |
0.058 |
| 262 |
250676_at |
AT5G06320
|
NHL3, NDR1/HIN1-like 3 |
0.182 |
-0.031 |
| 263 |
251192_at |
AT3G62720
|
ATXT1, XT1, xylosyltransferase 1, XXT1, XYG XYLOSYLTRANSFERASE 1 |
0.182 |
-0.019 |
| 264 |
254107_at |
AT4G25220
|
AtG3Pp2, glycerol-3-phosphate permease 2, G3Pp2, glycerol-3-phosphate permease 2, RHS15, root hair specific 15 |
0.182 |
-0.046 |
| 265 |
266072_at |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
0.181 |
0.167 |
| 266 |
257322_at |
ATMG01180
|
ORF111B |
0.181 |
0.065 |
| 267 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
0.181 |
0.035 |
| 268 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.181 |
0.059 |
| 269 |
249234_at |
AT5G42200
|
[RING/U-box superfamily protein] |
0.180 |
0.053 |
| 270 |
262333_at |
AT1G64020
|
[Serine protease inhibitor (SERPIN) family protein] |
0.180 |
0.152 |
| 271 |
254208_at |
AT4G24175
|
unknown |
0.180 |
-0.090 |
| 272 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.180 |
0.219 |
| 273 |
246396_at |
AT1G58180
|
ATBCA6, A. THALIANA BETA CARBONIC ANHYDRASE 6, BCA6, beta carbonic anhydrase 6 |
0.179 |
0.137 |
| 274 |
251641_at |
AT3G57470
|
[Insulinase (Peptidase family M16) family protein] |
0.179 |
0.008 |
| 275 |
264507_at |
AT1G09415
|
NIMIN-3, NIM1-interacting 3 |
0.178 |
0.125 |
| 276 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
0.178 |
0.122 |
| 277 |
247731_at |
AT5G59590
|
UGT76E2, UDP-glucosyl transferase 76E2 |
0.178 |
-0.016 |
| 278 |
259160_at |
AT3G05410
|
[Photosystem II reaction center PsbP family protein] |
0.178 |
-0.049 |
| 279 |
250868_at |
AT5G03860
|
MLS, malate synthase |
0.178 |
-0.091 |
| 280 |
266518_at |
AT2G35170
|
[Histone H3 K4-specific methyltransferase SET7/9 family protein] |
0.177 |
0.053 |
| 281 |
248563_at |
AT5G49690
|
[UDP-Glycosyltransferase superfamily protein] |
0.177 |
0.031 |
| 282 |
253485_at |
AT4G31800
|
ATWRKY18, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 18, WRKY18, WRKY DNA-binding protein 18 |
0.177 |
-0.097 |
| 283 |
254324_at |
AT4G22640
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.177 |
-0.073 |
| 284 |
245777_at |
AT1G73540
|
atnudt21, nudix hydrolase homolog 21, NUDT21, nudix hydrolase homolog 21 |
0.177 |
0.096 |
| 285 |
255931_at |
AT1G12710
|
AtPP2-A12, phloem protein 2-A12, PP2-A12, phloem protein 2-A12 |
0.177 |
0.014 |
| 286 |
245925_at |
AT5G28770
|
AtbZIP63, Arabidopsis thaliana basic leucine zipper 63, BZO2H3 |
0.177 |
0.132 |
| 287 |
249111_at |
AT5G43770
|
[proline-rich family protein] |
0.176 |
-0.011 |
| 288 |
265086_at |
AT1G03770
|
ATRING1B, ARABIDOPSIS THALIANA RING 1B, RING1B, RING 1B |
0.176 |
0.023 |
| 289 |
248199_at |
AT5G54170
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.176 |
0.005 |
| 290 |
248405_at |
AT5G51480
|
SKS2, SKU5 similar 2 |
0.175 |
0.020 |
| 291 |
262510_at |
AT1G11270
|
[F-box and associated interaction domains-containing protein] |
0.175 |
0.022 |
| 292 |
253829_at |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
0.175 |
0.140 |
| 293 |
264912_at |
AT1G60750
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.175 |
0.051 |
| 294 |
261938_at |
AT1G22510
|
[FUNCTIONS IN: zinc ion binding] |
0.175 |
-0.072 |
| 295 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
0.175 |
0.079 |
| 296 |
254027_at |
AT4G25835
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.175 |
-0.033 |
| 297 |
262635_at |
AT1G06570
|
HPD, 4-hydroxyphenylpyruvate dioxygenase, HPPD, 4-hydroxyphenylpyruvate dioxygenase, PDS1, phytoene desaturation 1 |
0.175 |
0.099 |
| 298 |
267300_at |
AT2G30140
|
UGT87A2, UDP-glucosyl transferase 87A2 |
0.174 |
0.123 |
| 299 |
249029_at |
AT5G44870
|
LAZ5, LAZARUS 5, TTR1, tolerance to Tobacco ringspot virus 1 |
0.174 |
0.041 |
| 300 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
0.174 |
-0.215 |
| 301 |
253707_at |
AT4G29200
|
[Beta-galactosidase related protein] |
-0.855 |
0.067 |
| 302 |
263098_at |
AT2G16005
|
[MD-2-related lipid recognition domain-containing protein] |
-0.840 |
-0.026 |
| 303 |
249545_at |
AT5G38030
|
[MATE efflux family protein] |
-0.809 |
-0.018 |
| 304 |
249567_at |
AT5G38020
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.750 |
0.047 |
| 305 |
248727_at |
AT5G47990
|
CYP705A5, cytochrome P450, family 705, subfamily A, polypeptide 5, THAD, THALIAN-DIOL DESATURASE, THAD1, THALIAN-DIOL DESATURASE |
-0.710 |
0.007 |
| 306 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
-0.676 |
0.079 |
| 307 |
247463_at |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
-0.669 |
-0.045 |
| 308 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
-0.652 |
-0.211 |
| 309 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.643 |
-0.033 |
| 310 |
248728_at |
AT5G48000
|
CYP708 A2, CYTOCHROME P450, FAMILY 708, SUBFAMILY A, POLYPEPTIDE 2, CYP708A2, cytochrome P450, family 708, subfamily A, polypeptide 2, THAH, THALIANOL HYDROXYLASE, THAH1, THALIANOL HYDROXYLASE 1 |
-0.634 |
0.019 |
| 311 |
264143_at |
AT1G79330
|
AMC6, ATMC5, metacaspase 5, ATMCP2b, metacaspase 2b, MC5, metacaspase 5, MCP2b, metacaspase 2b |
-0.589 |
-0.028 |
| 312 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
-0.584 |
0.000 |
| 313 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
-0.577 |
0.059 |
| 314 |
256631_at |
AT3G28320
|
unknown |
-0.571 |
-0.038 |
| 315 |
267317_at |
AT2G34700
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.560 |
-0.010 |
| 316 |
260805_at |
AT1G78320
|
ATGSTU23, glutathione S-transferase TAU 23, GSTU23, glutathione S-transferase TAU 23 |
-0.555 |
0.015 |
| 317 |
250764_at |
AT5G05960
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.550 |
0.008 |
| 318 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.546 |
-0.057 |
| 319 |
255284_at |
AT4G04610
|
APR, APR1, APS reductase 1, ATAPR1, PRH19, PAPS REDUCTASE HOMOLOG 19 |
-0.528 |
-0.015 |
| 320 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.527 |
0.043 |
| 321 |
259264_at |
AT3G01260
|
[Galactose mutarotase-like superfamily protein] |
-0.507 |
-0.081 |
| 322 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.503 |
-0.070 |
| 323 |
245736_at |
AT1G73330
|
ATDR4, drought-repressed 4, DR4, drought-repressed 4 |
-0.498 |
0.007 |
| 324 |
256878_at |
AT3G26460
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.491 |
0.030 |
| 325 |
250561_at |
AT5G08030
|
AtGDPD6, GDPD6, glycerophosphodiester phosphodiesterase 6 |
-0.472 |
0.011 |
| 326 |
266996_at |
AT2G34490
|
CYP710A2, cytochrome P450, family 710, subfamily A, polypeptide 2 |
-0.471 |
0.013 |
| 327 |
265122_at |
AT1G62540
|
FMO GS-OX2, flavin-monooxygenase glucosinolate S-oxygenase 2 |
-0.460 |
0.046 |
| 328 |
258367_at |
AT3G14370
|
WAG2 |
-0.453 |
-0.083 |
| 329 |
248729_at |
AT5G48010
|
AtTHAS1, Arabidopsis thaliana thalianol synthase 1, THAS, THALIANOL SYNTHASE, THAS1, thalianol synthase 1 |
-0.449 |
0.019 |
| 330 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
-0.434 |
-0.029 |
| 331 |
265327_at |
AT2G18210
|
unknown |
-0.426 |
-0.026 |
| 332 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
-0.423 |
-0.043 |
| 333 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.423 |
0.014 |
| 334 |
247549_at |
AT5G61420
|
AtMYB28, HAG1, HIGH ALIPHATIC GLUCOSINOLATE 1, MYB28, myb domain protein 28, PMG1, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 1 |
-0.421 |
-0.007 |
| 335 |
261660_at |
AT1G18370
|
ATNACK1, ARABIDOPSIS NPK1-ACTIVATING KINESIN 1, HIK, HINKEL, NACK1, NPK1-ACTIVATING KINESIN 1 |
-0.419 |
-0.026 |
| 336 |
254907_at |
AT4G11190
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.419 |
-0.007 |
| 337 |
248807_at |
AT5G47500
|
PME5, pectin methylesterase 5 |
-0.415 |
-0.010 |
| 338 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
-0.415 |
0.015 |
| 339 |
245047_at |
ATCG00020
|
PSBA, photosystem II reaction center protein A |
-0.413 |
0.046 |
| 340 |
252691_at |
AT3G44050
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.410 |
-0.067 |
| 341 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-0.406 |
-0.032 |
| 342 |
253254_at |
AT4G34650
|
SQS2, squalene synthase 2 |
-0.405 |
-0.001 |
| 343 |
254931_at |
AT4G11460
|
CRK30, cysteine-rich RLK (RECEPTOR-like protein kinase) 30 |
-0.398 |
-0.053 |
| 344 |
248963_at |
AT5G45700
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.391 |
-0.079 |
| 345 |
257793_at |
AT3G26960
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.384 |
-0.023 |
| 346 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
-0.383 |
0.087 |
| 347 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
-0.381 |
0.141 |
| 348 |
267457_at |
AT2G33790
|
AGP30, arabinogalactan protein 30, ATAGP30 |
-0.380 |
-0.075 |
| 349 |
247337_at |
AT5G63660
|
LCR74, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 74, PDF2.5 |
-0.379 |
-0.021 |
| 350 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
-0.378 |
-0.151 |
| 351 |
251299_at |
AT3G61950
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.376 |
0.018 |
| 352 |
256602_at |
AT3G28310
|
unknown |
-0.374 |
-0.065 |
| 353 |
254606_at |
AT4G19030
|
AT-NLM1, ATNLM1, NOD26-LIKE MAJOR INTRINSIC PROTEIN 1, NIP1;1, NOD26-LIKE INTRINSIC PROTEIN 1;1, NLM1, NOD26-like major intrinsic protein 1 |
-0.374 |
-0.060 |
| 354 |
264319_at |
AT1G04110
|
AtSDD1, SDD1, STOMATAL DENSITY AND DISTRIBUTION 1 |
-0.374 |
0.010 |
| 355 |
248413_at |
AT5G51600
|
ATMAP65-3, ARABIDOPSIS THALIANA MICROTUBULE-ASSOCIATED PROTEIN 65-3, MAP65-3, MICROTUBULE-ASSOCIATED PROTEIN 65-3, PLE, PLEIADE |
-0.373 |
0.099 |
| 356 |
256781_at |
AT3G13650
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.373 |
-0.036 |
| 357 |
256211_at |
AT1G50960
|
ATGA2OX7, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 7, GA2OX7, gibberellin 2-oxidase 7 |
-0.370 |
-0.026 |
| 358 |
263034_at |
AT1G24020
|
MLP423, MLP-like protein 423 |
-0.369 |
-0.017 |
| 359 |
246415_at |
AT5G17160
|
unknown |
-0.369 |
0.016 |
| 360 |
247577_at |
AT5G61290
|
[Flavin-binding monooxygenase family protein] |
-0.368 |
0.039 |
| 361 |
251058_at |
AT5G01790
|
unknown |
-0.365 |
-0.000 |
| 362 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
-0.365 |
0.053 |
| 363 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
-0.358 |
-0.021 |
| 364 |
253527_at |
AT4G31470
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.355 |
0.063 |
| 365 |
251509_at |
AT3G59010
|
PME35, pectin methylesterase 35, PME61, pectin methylesterase 61 |
-0.355 |
-0.032 |
| 366 |
253155_at |
AT4G35720
|
unknown |
-0.353 |
-0.078 |
| 367 |
260059_at |
AT1G78090
|
ATTPPB, Arabidopsis thaliana trehalose-6-phosphate phosphatase B, TPPB, trehalose-6-phosphate phosphatase B |
-0.350 |
-0.025 |
| 368 |
246557_at |
AT5G15510
|
[TPX2 (targeting protein for Xklp2) protein family] |
-0.349 |
-0.056 |
| 369 |
262162_at |
AT1G78020
|
unknown |
-0.348 |
-0.017 |
| 370 |
263318_at |
AT2G24762
|
AtGDU4, glutamine dumper 4, GDU4, glutamine dumper 4 |
-0.347 |
-0.036 |
| 371 |
256368_at |
AT1G66800
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.343 |
0.053 |
| 372 |
249599_at |
AT5G37990
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.342 |
-0.132 |
| 373 |
255460_at |
AT4G02800
|
unknown |
-0.341 |
-0.059 |
| 374 |
248057_at |
AT5G55520
|
unknown |
-0.339 |
-0.007 |
| 375 |
256489_at |
AT1G31550
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.338 |
-0.010 |
| 376 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
-0.337 |
0.019 |
| 377 |
246920_at |
AT5G25090
|
AtENODL13, ENODL13, early nodulin-like protein 13 |
-0.337 |
-0.021 |
| 378 |
258017_at |
AT3G19370
|
unknown |
-0.335 |
-0.072 |
| 379 |
256096_at |
AT1G13650
|
unknown |
-0.334 |
-0.111 |
| 380 |
248888_at |
AT5G46240
|
AtKAT1, KAT1, potassium channel in Arabidopsis thaliana 1 |
-0.333 |
0.027 |
| 381 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
-0.333 |
-0.071 |
| 382 |
255856_at |
AT1G66940
|
[protein kinase-related] |
-0.332 |
0.033 |
| 383 |
255265_at |
AT4G05190
|
ATK5, kinesin 5 |
-0.329 |
-0.004 |
| 384 |
250661_at |
AT5G07030
|
[Eukaryotic aspartyl protease family protein] |
-0.329 |
-0.002 |
| 385 |
264647_at |
AT1G09090
|
ATRBOHB, respiratory burst oxidase homolog B, ATRBOHB-BETA, RBOHB, respiratory burst oxidase homolog B |
-0.328 |
-0.030 |
| 386 |
251298_at |
AT3G62040
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.328 |
-0.015 |
| 387 |
254098_at |
AT4G25100
|
ATFSD1, ARABIDOPSIS FE SUPEROXIDE DISMUTASE 1, FSD1, Fe superoxide dismutase 1 |
-0.328 |
-0.001 |
| 388 |
259549_at |
AT1G35290
|
ALT1, acyl-lipid thioesterase 1 |
-0.327 |
-0.012 |
| 389 |
263498_at |
AT2G42610
|
LSH10, LIGHT SENSITIVE HYPOCOTYLS 10 |
-0.327 |
-0.032 |
| 390 |
255957_at |
AT1G22160
|
unknown |
-0.325 |
0.021 |
| 391 |
257115_at |
AT3G20150
|
[Kinesin motor family protein] |
-0.323 |
-0.024 |
| 392 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
-0.322 |
-0.012 |
| 393 |
261449_at |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
-0.320 |
-0.007 |
| 394 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-0.318 |
-0.187 |
| 395 |
245739_at |
AT1G44110
|
CYCA1;1, Cyclin A1;1 |
-0.317 |
-0.029 |
| 396 |
261368_at |
AT1G53070
|
[Legume lectin family protein] |
-0.317 |
-0.040 |
| 397 |
257774_at |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
-0.316 |
-0.042 |
| 398 |
253051_at |
AT4G37490
|
CYC1, CYCLIN 1, CYCB1, CYCB1;1, CYCLIN B1;1 |
-0.316 |
-0.074 |
| 399 |
266465_at |
AT2G47750
|
GH3.9, putative indole-3-acetic acid-amido synthetase GH3.9 |
-0.315 |
0.011 |
| 400 |
252983_at |
AT4G37980
|
ATCAD7, CAD7, CINNAMYL-ALCOHOL DEHYDROGENASE 7, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-1, elicitor-activated gene 3-1 |
-0.311 |
0.027 |
| 401 |
264404_at |
AT2G25160
|
CYP82F1, cytochrome P450, family 82, subfamily F, polypeptide 1 |
-0.311 |
0.018 |
| 402 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
-0.306 |
-0.015 |
| 403 |
255450_at |
AT4G02850
|
[phenazine biosynthesis PhzC/PhzF family protein] |
-0.305 |
-0.018 |
| 404 |
249848_at |
AT5G23220
|
NIC3, nicotinamidase 3 |
-0.305 |
-0.020 |
| 405 |
259985_at |
AT1G76620
|
unknown |
-0.305 |
-0.011 |
| 406 |
262081_at |
AT1G59540
|
ZCF125 |
-0.302 |
0.033 |
| 407 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.302 |
0.117 |
| 408 |
248619_at |
AT5G49630
|
AAP6, amino acid permease 6 |
-0.300 |
-0.009 |
| 409 |
261921_at |
AT1G65900
|
unknown |
-0.299 |
-0.048 |
| 410 |
247880_at |
AT5G57780
|
P1R1, P1R1 |
-0.299 |
0.166 |
| 411 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.299 |
-0.023 |
| 412 |
256062_at |
AT1G07090
|
LSH6, LIGHT SENSITIVE HYPOCOTYLS 6 |
-0.299 |
-0.007 |
| 413 |
252530_at |
AT3G46500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.298 |
-0.000 |
| 414 |
251746_at |
AT3G56060
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
-0.297 |
0.100 |
| 415 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
-0.294 |
-0.066 |
| 416 |
267636_at |
AT2G42110
|
unknown |
-0.294 |
-0.079 |
| 417 |
248725_at |
AT5G47980
|
[HXXXD-type acyl-transferase family protein] |
-0.291 |
-0.077 |
| 418 |
247603_at |
AT5G60930
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.290 |
-0.065 |
| 419 |
259736_at |
AT1G64390
|
AtGH9C2, glycosyl hydrolase 9C2, GH9C2, glycosyl hydrolase 9C2 |
-0.289 |
-0.060 |
| 420 |
259813_at |
AT1G49860
|
ATGSTF14, glutathione S-transferase (class phi) 14, GSTF14, glutathione S-transferase (class phi) 14 |
-0.289 |
-0.315 |
| 421 |
264481_at |
AT1G77200
|
[Integrase-type DNA-binding superfamily protein] |
-0.288 |
0.022 |
| 422 |
248625_at |
AT5G48880
|
KAT5, 3-KETO-ACYL-COENZYME A THIOLASE 5, PKT1, PEROXISOMAL-3-KETO-ACYL-COA THIOLASE 1, PKT2, peroxisomal 3-keto-acyl-CoA thiolase 2 |
-0.288 |
-0.059 |
| 423 |
249814_at |
AT5G23840
|
[MD-2-related lipid recognition domain-containing protein] |
-0.287 |
-0.205 |
| 424 |
262109_at |
AT1G02730
|
ATCSLD5, cellulose synthase-like D5, CSLD5, CELLULOSE SYNTHASE LIKE D5, CSLD5, cellulose synthase-like D5, SOS6, SALT OVERLY SENSITIVE 6 |
-0.287 |
-0.003 |
| 425 |
258470_at |
AT3G06035
|
[Glycoprotein membrane precursor GPI-anchored] |
-0.286 |
-0.028 |
| 426 |
257740_at |
AT3G27330
|
[zinc finger (C3HC4-type RING finger) family protein] |
-0.286 |
-0.005 |
| 427 |
256128_at |
AT1G18140
|
ATLAC1, LAC1, laccase 1 |
-0.284 |
0.045 |
| 428 |
256349_at |
AT1G54890
|
[Late embryogenesis abundant (LEA) protein-related] |
-0.284 |
-0.146 |
| 429 |
256599_at |
AT3G14760
|
unknown |
-0.283 |
0.038 |
| 430 |
252148_at |
AT3G51280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.283 |
-0.059 |
| 431 |
259930_at |
AT1G34355
|
ATPS1, PARALLEL SPINDLE 1, PS1, PARALLEL SPINDLE 1 |
-0.280 |
0.043 |
| 432 |
255852_at |
AT1G66970
|
GDPDL1, Glycerophosphodiester phosphodiesterase (GDPD) like 1, SVL2, SHV3-like 2 |
-0.279 |
0.027 |
| 433 |
265085_at |
AT1G03780
|
AtTPX2, TPX2, targeting protein for XKLP2 |
-0.278 |
0.025 |
| 434 |
251067_at |
AT5G01910
|
unknown |
-0.278 |
-0.112 |
| 435 |
246247_at |
AT4G36640
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.278 |
-0.031 |
| 436 |
252149_at |
AT3G51290
|
APSR1, Altered Phosphate Starvation Response 1 |
-0.276 |
-0.036 |
| 437 |
258464_at |
AT3G17360
|
POK1, phragmoplast orienting kinesin 1 |
-0.276 |
-0.000 |
| 438 |
263715_at |
AT2G20570
|
GBF's pro-rich region-interacting factor 1 |
-0.276 |
-0.023 |
| 439 |
261804_at |
AT1G30530
|
UGT78D1, UDP-glucosyl transferase 78D1 |
-0.274 |
-0.129 |
| 440 |
261768_at |
AT1G15550
|
ATGA3OX1, ARABIDOPSIS THALIANA GIBBERELLIN 3 BETA-HYDROXYLASE 1, GA3OX1, gibberellin 3-oxidase 1, GA4, GA REQUIRING 4 |
-0.274 |
-0.033 |
| 441 |
250228_at |
AT5G13840
|
CCS52B, cell cycle switch protein 52 B, FZR3, FIZZY-related 3 |
-0.273 |
0.001 |
| 442 |
266669_at |
AT2G29750
|
UGT71C1, UDP-glucosyl transferase 71C1 |
-0.272 |
-0.005 |
| 443 |
258003_at |
AT3G29030
|
ATEXP5, ARABIDOPSIS THALIANA EXPANSIN 5, ATEXPA5, ARABIDOPSIS THALIANA EXPANSIN A5, ATHEXP ALPHA 1.4, EXP5, EXPANSIN 5, EXPA5, expansin A5 |
-0.271 |
-0.020 |
| 444 |
262659_at |
AT1G14240
|
[GDA1/CD39 nucleoside phosphatase family protein] |
-0.271 |
-0.007 |
| 445 |
262646_at |
AT1G62800
|
ASP4, aspartate aminotransferase 4 |
-0.270 |
-0.034 |
| 446 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
-0.270 |
0.049 |
| 447 |
252916_at |
AT4G38950
|
[ATP binding microtubule motor family protein] |
-0.270 |
-0.055 |
| 448 |
248961_at |
AT5G45650
|
[subtilase family protein] |
-0.270 |
-0.102 |
| 449 |
245343_at |
AT4G15830
|
[ARM repeat superfamily protein] |
-0.270 |
-0.051 |
| 450 |
258098_at |
AT3G23670
|
KINESIN-12B, PAKRP1L |
-0.269 |
-0.026 |
| 451 |
259403_at |
AT1G17745
|
PGDH, 3-phosphoglycerate dehydrogenase, PGDH2, phosphoglycerate dehydrogenase 2 |
-0.268 |
0.003 |
| 452 |
256673_at |
AT3G52370
|
FLA15, FASCICLIN-like arabinogalactan protein 15 precursor |
-0.268 |
-0.066 |
| 453 |
260565_at |
AT2G43800
|
[Actin-binding FH2 (formin homology 2) family protein] |
-0.268 |
-0.046 |
| 454 |
260146_at |
AT1G52770
|
[Phototropic-responsive NPH3 family protein] |
-0.266 |
0.024 |
| 455 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
-0.265 |
-0.162 |
| 456 |
251287_at |
AT3G61820
|
[Eukaryotic aspartyl protease family protein] |
-0.264 |
-0.039 |
| 457 |
248540_at |
AT5G50160
|
ATFRO8, FERRIC REDUCTION OXIDASE 8, FRO8, ferric reduction oxidase 8 |
-0.263 |
0.051 |
| 458 |
253650_at |
AT4G30020
|
[PA-domain containing subtilase family protein] |
-0.263 |
-0.042 |
| 459 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
-0.262 |
-0.020 |
| 460 |
253788_at |
AT4G28680
|
AtTYDC, TYDC, L-tyrosine decarboxylase, TYRDC, L-tyrosine decarboxylase, TYRDC1, L-TYROSINE DECARBOXYLASE 1 |
-0.262 |
0.027 |
| 461 |
248309_at |
AT5G52540
|
unknown |
-0.261 |
-0.040 |
| 462 |
258270_at |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
-0.260 |
-0.120 |
| 463 |
264341_at |
AT1G70270
|
unknown |
-0.260 |
-0.001 |
| 464 |
266089_at |
AT2G38010
|
[Neutral/alkaline non-lysosomal ceramidase] |
-0.260 |
0.002 |
| 465 |
253024_at |
AT4G38080
|
[hydroxyproline-rich glycoprotein family protein] |
-0.260 |
0.106 |
| 466 |
267618_at |
AT2G26760
|
CYCB1;4, Cyclin B1;4 |
-0.260 |
-0.019 |
| 467 |
246585_at |
AT5G14750
|
ATMYB66, myb domain protein 66, MYB66, myb domain protein 66, WER, WEREWOLF, WER1, WEREWOLF 1 |
-0.259 |
0.087 |
| 468 |
251368_at |
AT3G61380
|
TRM14, TON1 Recruiting Motif 14 |
-0.258 |
-0.009 |
| 469 |
266784_at |
AT2G28960
|
[Leucine-rich repeat protein kinase family protein] |
-0.257 |
-0.035 |
| 470 |
265261_at |
AT2G42990
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.257 |
0.047 |
| 471 |
261177_at |
AT1G04770
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.257 |
-0.082 |
| 472 |
265545_at |
AT2G28250
|
NCRK |
-0.256 |
-0.027 |
| 473 |
262819_at |
AT1G11600
|
CYP77B1, cytochrome P450, family 77, subfamily B, polypeptide 1 |
-0.255 |
-0.006 |
| 474 |
261772_at |
AT1G76240
|
unknown |
-0.255 |
0.020 |
| 475 |
254233_at |
AT4G23800
|
3xHMG-box2, 3xHigh Mobility Group-box2 |
-0.254 |
-0.049 |
| 476 |
253804_at |
AT4G28230
|
unknown |
-0.254 |
0.021 |
| 477 |
246002_at |
AT5G20740
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.253 |
-0.080 |
| 478 |
258376_at |
AT3G17680
|
[Kinase interacting (KIP1-like) family protein] |
-0.253 |
-0.019 |
| 479 |
259851_at |
AT1G72250
|
[Di-glucose binding protein with Kinesin motor domain] |
-0.251 |
-0.024 |
| 480 |
264403_at |
AT2G25150
|
[HXXXD-type acyl-transferase family protein] |
-0.250 |
-0.038 |
| 481 |
249809_at |
AT5G23910
|
[ATP binding microtubule motor family protein] |
-0.249 |
0.062 |
| 482 |
267078_at |
AT2G40960
|
[Single-stranded nucleic acid binding R3H protein] |
-0.248 |
-0.094 |
| 483 |
264071_at |
AT2G27920
|
SCPL51, serine carboxypeptidase-like 51 |
-0.248 |
-0.109 |
| 484 |
264745_at |
AT1G62180
|
5'adenylylphosphosulfate reductase 2 |
-0.248 |
-0.025 |
| 485 |
246986_at |
AT5G67280
|
RLK, receptor-like kinase |
-0.248 |
-0.049 |
| 486 |
261780_at |
AT1G76310
|
CYCB2;4, CYCLIN B2;4 |
-0.248 |
0.005 |
| 487 |
264271_at |
AT1G60270
|
BGLU6, beta glucosidase 6 |
-0.248 |
-0.053 |
| 488 |
254361_at |
AT4G22212
|
[Arabidopsis defensin-like protein] |
-0.247 |
-0.073 |
| 489 |
256864_at |
AT3G23890
|
ATTOPII, TOPII, topoisomerase II |
-0.246 |
-0.043 |
| 490 |
249894_at |
AT5G22580
|
[Stress responsive A/B Barrel Domain] |
-0.245 |
0.012 |
| 491 |
256721_at |
AT2G34150
|
ATRANGAP2, ATSCAR1, SCAR1, WAVE1, WISKOTT-ALDRICH SYNDROME PROTEIN FAMILY VERPROLIN HOMOLOGOUS PROTEIN 1 |
-0.244 |
0.015 |
| 492 |
256125_at |
AT1G18250
|
ATLP-1 |
-0.243 |
-0.011 |
| 493 |
262811_at |
AT1G11700
|
unknown |
-0.243 |
-0.120 |
| 494 |
247039_at |
AT5G67270
|
ATEB1C, MICROTUBULE END BINDING PROTEIN 1, ATEB1H1, ATEB1-HOMOLOG1, EB1C, end binding protein 1C |
-0.242 |
-0.026 |
| 495 |
259481_at |
AT1G18970
|
GLP4, germin-like protein 4 |
-0.241 |
0.047 |
| 496 |
264511_at |
AT1G09350
|
AtGolS3, galactinol synthase 3, GolS3, galactinol synthase 3 |
-0.240 |
0.032 |
| 497 |
260812_at |
AT1G43650
|
UMAMIT22, Usually multiple acids move in and out Transporters 22 |
-0.239 |
-0.029 |
| 498 |
249675_at |
AT5G35940
|
[Mannose-binding lectin superfamily protein] |
-0.239 |
-0.161 |
| 499 |
250045_at |
AT5G17700
|
[MATE efflux family protein] |
-0.238 |
0.032 |
| 500 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
-0.238 |
-0.008 |
| 501 |
257875_at |
AT3G17120
|
unknown |
-0.238 |
0.001 |
| 502 |
252911_at |
AT4G39510
|
CYP96A12, cytochrome P450, family 96, subfamily A, polypeptide 12 |
-0.238 |
-0.005 |
| 503 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
-0.238 |
-0.047 |
| 504 |
249970_at |
AT5G19100
|
[Eukaryotic aspartyl protease family protein] |
-0.236 |
0.018 |
| 505 |
254235_at |
AT4G23750
|
CRF2, cytokinin response factor 2, TMO3, TARGET OF MONOPTEROS 3 |
-0.236 |
-0.081 |
| 506 |
261431_at |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
-0.236 |
0.150 |
| 507 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
-0.236 |
-0.041 |
| 508 |
245259_at |
AT4G14150
|
KINESIN-12A, PAKRP1, phragmoplast-associated kinesin-related protein 1 |
-0.236 |
0.103 |
| 509 |
254909_at |
AT4G11210
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.235 |
-0.021 |
| 510 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
-0.234 |
-0.008 |
| 511 |
255403_at |
AT4G03400
|
DFL2, DWARF IN LIGHT 2, GH3-10 |
-0.234 |
0.026 |
| 512 |
263942_at |
AT2G35860
|
FLA16, FASCICLIN-like arabinogalactan protein 16 precursor |
-0.234 |
-0.073 |
| 513 |
262752_at |
AT1G16330
|
CYCB3;1, cyclin b3;1 |
-0.234 |
-0.004 |
| 514 |
252414_at |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
-0.233 |
-0.022 |
| 515 |
256145_at |
AT1G48750
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.233 |
0.051 |
| 516 |
260297_at |
AT1G80280
|
[alpha/beta-Hydrolases superfamily protein] |
-0.233 |
0.007 |
| 517 |
262508_at |
AT1G11300
|
[protein serine/threonine kinases] |
-0.233 |
0.006 |
| 518 |
266738_at |
AT2G47010
|
unknown |
-0.231 |
-0.030 |
| 519 |
263016_at |
AT1G23410
|
[Ribosomal protein S27a / Ubiquitin family protein] |
-0.230 |
-0.005 |
| 520 |
259656_at |
AT1G55200
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.230 |
-0.019 |
| 521 |
249347_at |
AT5G40830
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.229 |
-0.101 |
| 522 |
251113_at |
AT5G01370
|
ACI1, ALC-interacting protein 1, TRM29, TON1 Recruiting Motif 29 |
-0.229 |
0.025 |
| 523 |
247354_at |
AT5G63590
|
ATFLS3, FLS3, flavonol synthase 3 |
-0.229 |
0.054 |
| 524 |
264091_at |
AT1G79110
|
BRG2, BOI-related gene 2 |
-0.227 |
-0.087 |
| 525 |
245584_at |
AT4G14940
|
AO1, amine oxidase 1, ATAO1, amine oxidase 1 |
-0.227 |
0.023 |
| 526 |
266912_at |
AT2G45900
|
TRM13, TON1 Recruiting Motif 13 |
-0.227 |
-0.121 |
| 527 |
255345_at |
AT4G04460
|
[Saposin-like aspartyl protease family protein] |
-0.226 |
-0.007 |
| 528 |
267010_at |
AT2G39250
|
SNZ, SCHNARCHZAPFEN |
-0.226 |
-0.076 |
| 529 |
246368_at |
AT1G51890
|
[Leucine-rich repeat protein kinase family protein] |
-0.224 |
0.015 |
| 530 |
257663_at |
AT3G20260
|
[FUNCTIONS IN: structural constituent of ribosome] |
-0.224 |
0.020 |
| 531 |
260363_at |
AT1G70550
|
unknown |
-0.223 |
0.020 |
| 532 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.223 |
0.040 |
| 533 |
247441_at |
AT5G62710
|
[Leucine-rich repeat protein kinase family protein] |
-0.223 |
-0.024 |
| 534 |
262758_at |
AT1G10780
|
[F-box/RNI-like superfamily protein] |
-0.223 |
-0.056 |
| 535 |
259365_at |
AT1G13300
|
HRS1, HYPERSENSITIVITY TO LOW PI-ELICITED PRIMARY ROOT SHORTENING 1 |
-0.222 |
-0.025 |
| 536 |
249167_at |
AT5G42860
|
unknown |
-0.222 |
-0.056 |
| 537 |
247827_at |
AT5G58500
|
LSH5, LIGHT SENSITIVE HYPOCOTYLS 5 |
-0.222 |
0.097 |
| 538 |
246021_at |
AT5G21100
|
[Plant L-ascorbate oxidase] |
-0.221 |
0.008 |
| 539 |
245341_at |
AT4G16447
|
unknown |
-0.220 |
-0.038 |
| 540 |
265405_at |
AT2G16750
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.220 |
-0.007 |
| 541 |
260693_at |
AT1G32450
|
AtNPF7.3, NPF7.3, NRT1/ PTR family 7.3, NRT1.5, nitrate transporter 1.5 |
-0.219 |
0.032 |
| 542 |
247755_at |
AT5G59090
|
ATSBT4.12, subtilase 4.12, SBT4.12, subtilase 4.12 |
-0.218 |
-0.084 |
| 543 |
254663_at |
AT4G18290
|
KAT2, potassium channel in Arabidopsis thaliana 2 |
-0.218 |
0.059 |
| 544 |
254371_at |
AT4G21760
|
BGLU47, beta-glucosidase 47 |
-0.218 |
-0.101 |
| 545 |
259912_at |
AT1G72670
|
iqd8, IQ-domain 8 |
-0.216 |
-0.039 |
| 546 |
256926_at |
AT3G22540
|
unknown |
-0.216 |
-0.088 |
| 547 |
250601_at |
AT5G07810
|
[SNF2 domain-containing protein / helicase domain-containing protein / HNH endonuclease domain-containing protein] |
-0.216 |
-0.013 |
| 548 |
248613_at |
AT5G49555
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
-0.214 |
0.019 |
| 549 |
261399_at |
AT1G79620
|
[Leucine-rich repeat protein kinase family protein] |
-0.214 |
0.024 |
| 550 |
253483_at |
AT4G31910
|
BAT1, BR-related AcylTransferase1, PIZ, PIZZA |
-0.214 |
0.042 |
| 551 |
246108_at |
AT5G28630
|
[glycine-rich protein] |
-0.213 |
-0.031 |
| 552 |
249644_at |
AT5G37010
|
unknown |
-0.213 |
0.020 |
| 553 |
253052_at |
AT4G37310
|
CYP81H1, cytochrome P450, family 81, subfamily H, polypeptide 1 |
-0.213 |
0.068 |
| 554 |
267076_at |
AT2G41090
|
[Calcium-binding EF-hand family protein] |
-0.212 |
-0.041 |
| 555 |
266418_at |
AT2G38750
|
ANNAT4, annexin 4, AtANN4 |
-0.212 |
0.002 |
| 556 |
254343_at |
AT4G21990
|
APR3, APS reductase 3, ATAPR3, PRH-26, PRH26, PAPS REDUCTASE HOMOLOG 26 |
-0.212 |
-0.024 |
| 557 |
264802_at |
AT1G08560
|
ATSYP111, KN, KNOLLE, SYP111, syntaxin of plants 111 |
-0.211 |
0.002 |
| 558 |
262304_at |
AT1G70890
|
MLP43, MLP-like protein 43 |
-0.211 |
-0.042 |
| 559 |
253985_at |
AT4G26220
|
CCoAOMT7, caffeoyl coenzyme A ester O-methyltransferase 7 |
-0.211 |
-0.115 |
| 560 |
259290_at |
AT3G11520
|
CYC2, CYCLIN 2, CYCB1;3, CYCLIN B1;3 |
-0.211 |
0.048 |
| 561 |
247134_at |
AT5G66230
|
[Chalcone-flavanone isomerase family protein] |
-0.211 |
-0.047 |
| 562 |
250379_at |
AT5G11590
|
TINY2, TINY2 |
-0.210 |
-0.042 |
| 563 |
256675_at |
AT3G52170
|
[DNA binding] |
-0.210 |
-0.183 |
| 564 |
246933_at |
AT5G25160
|
ZFP3, zinc finger protein 3 |
-0.209 |
-0.029 |
| 565 |
267382_at |
AT2G44300
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.209 |
-0.015 |
| 566 |
259632_at |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
-0.209 |
-0.049 |
| 567 |
250418_at |
AT5G11240
|
[transducin family protein / WD-40 repeat family protein] |
-0.208 |
-0.142 |
| 568 |
258471_at |
AT3G06030
|
ANP3, NPK1-related protein kinase 3, AtANP3, MAPKKK12, NP3, NPK1-related protein kinase 3 |
-0.208 |
-0.043 |
| 569 |
252437_at |
AT3G47380
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.208 |
-0.021 |
| 570 |
261653_at |
AT1G01900
|
ATSBT1.1, SBTI1.1 |
-0.207 |
0.006 |
| 571 |
260331_at |
AT1G80270
|
PPR596, PENTATRICOPEPTIDE REPEAT 596 |
-0.206 |
-0.075 |
| 572 |
261878_at |
AT1G50560
|
CYP705A25, cytochrome P450, family 705, subfamily A, polypeptide 25 |
-0.206 |
0.043 |
| 573 |
247685_at |
AT5G59680
|
[Leucine-rich repeat protein kinase family protein] |
-0.205 |
-0.011 |
| 574 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.205 |
-0.028 |
| 575 |
246827_at |
AT5G26330
|
[Cupredoxin superfamily protein] |
-0.205 |
0.023 |
| 576 |
250277_at |
AT5G12940
|
[Leucine-rich repeat (LRR) family protein] |
-0.205 |
-0.002 |
| 577 |
258480_at |
AT3G02640
|
unknown |
-0.205 |
-0.001 |
| 578 |
252763_at |
AT3G42725
|
[Putative membrane lipoprotein] |
-0.204 |
0.081 |
| 579 |
256676_at |
AT3G52180
|
ATPTPKIS1, ATSEX4, DSP4, DUAL-SPECIFICITY PROTEIN PHOSPHATASE 4, SEX4, STARCH-EXCESS 4 |
-0.204 |
-0.153 |
| 580 |
258897_at |
AT3G05730
|
[LOCATED IN: endomembrane system] |
-0.204 |
0.023 |
| 581 |
266532_at |
AT2G16890
|
[UDP-Glycosyltransferase superfamily protein] |
-0.204 |
0.032 |
| 582 |
261636_at |
AT1G50110
|
[D-aminoacid aminotransferase-like PLP-dependent enzymes superfamily protein] |
-0.204 |
-0.115 |
| 583 |
251800_at |
AT3G55510
|
RBL, REBELOTE |
-0.203 |
0.003 |
| 584 |
262969_at |
AT1G75710
|
[C2H2-like zinc finger protein] |
-0.203 |
0.030 |
| 585 |
258867_at |
AT3G03130
|
unknown |
-0.203 |
-0.050 |
| 586 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
-0.202 |
-0.018 |
| 587 |
266640_at |
AT2G35585
|
unknown |
-0.202 |
-0.076 |
| 588 |
265326_at |
AT2G18220
|
[Noc2p family] |
-0.202 |
-0.121 |
| 589 |
256736_at |
AT3G29410
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.201 |
0.079 |
| 590 |
255648_at |
AT4G00910
|
[Aluminium activated malate transporter family protein] |
-0.201 |
0.035 |
| 591 |
261106_at |
AT1G62990
|
IXR11, KNAT7, KNOTTED-like homeobox of Arabidopsis thaliana 7 |
-0.201 |
-0.031 |
| 592 |
247962_at |
AT5G56580
|
ANQ1, ARABIDOPSIS NQK1, ATMKK6, ARABIDOPSIS THALIANA MAP KINASE KINASE 6, MKK6, MAP kinase kinase 6 |
-0.200 |
-0.051 |
| 593 |
249812_at |
AT5G23830
|
[MD-2-related lipid recognition domain-containing protein] |
-0.200 |
-0.045 |
| 594 |
264000_at |
AT2G22500
|
ATPUMP5, PLANT UNCOUPLING MITOCHONDRIAL PROTEIN 5, DIC1, DICARBOXYLATE CARRIER 1, UCP5, uncoupling protein 5 |
-0.199 |
-0.062 |
| 595 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
-0.199 |
-0.294 |
| 596 |
261907_at |
AT1G65060
|
4CL3, 4-coumarate:CoA ligase 3 |
-0.198 |
-0.101 |
| 597 |
255825_at |
AT2G40475
|
ASG8, ALTERED SEED GERMINATION 8 |
-0.198 |
0.011 |
| 598 |
256626_at |
AT3G20015
|
[Eukaryotic aspartyl protease family protein] |
-0.197 |
0.002 |
| 599 |
258505_at |
AT3G06530
|
[ARM repeat superfamily protein] |
-0.197 |
-0.114 |
| 600 |
251388_at |
AT3G60840
|
MAP65-4, microtubule-associated protein 65-4 |
-0.197 |
0.012 |