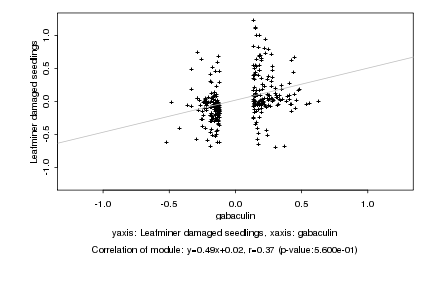
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
gabaculin |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
267121_at |
AT2G23540
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.622 |
0.012 |
| 2 |
250515_at |
AT5G09570
|
[Cox19-like CHCH family protein] |
0.555 |
-0.027 |
| 3 |
253024_at |
AT4G38080
|
[hydroxyproline-rich glycoprotein family protein] |
0.535 |
-0.040 |
| 4 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
0.478 |
0.186 |
| 5 |
250500_at |
AT5G09530
|
PELPK1, Pro-Glu-Leu|Ile|Val-Pro-Lys 1, PRP10, proline-rich protein 10 |
0.476 |
0.178 |
| 6 |
250541_at |
AT5G09520
|
PELPK2, Pro-Glu-Leu|Ile|Val-Pro-Lys 2 |
0.457 |
0.025 |
| 7 |
245889_at |
AT5G09480
|
[hydroxyproline-rich glycoprotein family protein] |
0.448 |
-0.091 |
| 8 |
245181_at |
AT5G12420
|
[O-acyltransferase (WSD1-like) family protein] |
0.446 |
0.672 |
| 9 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.443 |
0.120 |
| 10 |
264774_at |
AT1G22890
|
unknown |
0.435 |
0.444 |
| 11 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.423 |
-0.142 |
| 12 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.420 |
0.623 |
| 13 |
247765_at |
AT5G58860
|
CYP86, CYP86A1, cytochrome P450, family 86, subfamily A, polypeptide 1, HORST, hydroxylase of root suberized tissue |
0.416 |
-0.037 |
| 14 |
260234_at |
AT1G74460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.412 |
-0.036 |
| 15 |
260035_at |
AT1G68850
|
[Peroxidase superfamily protein] |
0.404 |
0.034 |
| 16 |
246464_at |
AT5G16980
|
[Zinc-binding dehydrogenase family protein] |
0.404 |
0.285 |
| 17 |
252209_at |
AT3G50400
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.401 |
0.096 |
| 18 |
260522_x_at |
AT2G41730
|
unknown |
0.393 |
0.082 |
| 19 |
265197_at |
AT2G36750
|
UGT73C1, UDP-glucosyl transferase 73C1 |
0.384 |
0.003 |
| 20 |
259432_at |
AT1G01520
|
ASG4, ALTERED SEED GERMINATION 4 |
0.370 |
-0.678 |
| 21 |
246203_at |
AT4G36610
|
[alpha/beta-Hydrolases superfamily protein] |
0.349 |
0.033 |
| 22 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.346 |
0.251 |
| 23 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
0.342 |
0.194 |
| 24 |
263998_at |
AT2G22510
|
[hydroxyproline-rich glycoprotein family protein] |
0.340 |
0.020 |
| 25 |
250230_at |
AT5G13900
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.328 |
0.033 |
| 26 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
0.328 |
0.097 |
| 27 |
262373_at |
AT1G73120
|
unknown |
0.324 |
-0.043 |
| 28 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.315 |
0.063 |
| 29 |
258905_at |
AT3G06390
|
[Uncharacterised protein family (UPF0497)] |
0.313 |
-0.053 |
| 30 |
250239_at |
AT5G13580
|
ABCG6, ATP-binding cassette G6 |
0.310 |
0.067 |
| 31 |
263005_at |
AT1G54540
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.304 |
0.103 |
| 32 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.300 |
0.207 |
| 33 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
0.300 |
-0.687 |
| 34 |
263515_at |
AT2G21640
|
unknown |
0.299 |
0.136 |
| 35 |
254474_at |
AT4G20390
|
[Uncharacterised protein family (UPF0497)] |
0.284 |
0.027 |
| 36 |
260582_at |
AT2G47200
|
unknown |
0.278 |
0.032 |
| 37 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
0.278 |
0.535 |
| 38 |
245543_at |
AT4G15260
|
[UDP-Glycosyltransferase superfamily protein] |
0.277 |
0.473 |
| 39 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
0.277 |
0.370 |
| 40 |
264403_at |
AT2G25150
|
[HXXXD-type acyl-transferase family protein] |
0.276 |
0.005 |
| 41 |
259282_at |
AT3G11430
|
ATGPAT5, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 5, GPAT5, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 5 |
0.275 |
0.031 |
| 42 |
250182_at |
AT5G14470
|
AtGALK2, GALK2, galactokinase 2 |
0.274 |
0.039 |
| 43 |
257670_at |
AT3G20340
|
unknown |
0.272 |
0.311 |
| 44 |
262406_at |
AT1G34670
|
AtMYB93, myb domain protein 93, MYB93, myb domain protein 93 |
0.268 |
0.053 |
| 45 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.268 |
0.717 |
| 46 |
256527_at |
AT1G66100
|
[Plant thionin] |
0.265 |
0.238 |
| 47 |
253496_at |
AT4G31870
|
ATGPX7, GLUTATHIONE PEROXIDASE 7, GPX7, glutathione peroxidase 7 |
0.264 |
-0.067 |
| 48 |
263825_at |
AT2G40370
|
LAC5, laccase 5 |
0.257 |
0.046 |
| 49 |
257666_at |
AT3G20270
|
[lipid-binding serum glycoprotein family protein] |
0.250 |
0.102 |
| 50 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
0.249 |
-0.050 |
| 51 |
250054_at |
AT5G17860
|
CAX7, calcium exchanger 7 |
0.248 |
0.383 |
| 52 |
254634_at |
AT4G18650
|
[transcription factor-related] |
0.247 |
0.019 |
| 53 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
0.245 |
0.792 |
| 54 |
244950_at |
ATMG00160
|
COX2, cytochrome oxidase 2 |
0.244 |
0.099 |
| 55 |
245352_at |
AT4G15490
|
UGT84A3 |
0.242 |
0.344 |
| 56 |
249364_at |
AT5G40590
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.240 |
0.056 |
| 57 |
264752_at |
AT1G23010
|
LPR1, Low Phosphate Root1 |
0.239 |
0.117 |
| 58 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
0.235 |
-0.505 |
| 59 |
249061_at |
AT5G44550
|
[Uncharacterised protein family (UPF0497)] |
0.233 |
-0.079 |
| 60 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
0.232 |
-0.425 |
| 61 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
0.226 |
-0.063 |
| 62 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.223 |
0.951 |
| 63 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.221 |
0.732 |
| 64 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.219 |
0.806 |
| 65 |
256217_at |
AT1G56320
|
unknown |
0.219 |
-0.031 |
| 66 |
255264_at |
AT4G05170
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.217 |
0.046 |
| 67 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
0.215 |
0.238 |
| 68 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
0.212 |
0.236 |
| 69 |
254404_at |
AT4G21340
|
B70 |
0.209 |
-0.008 |
| 70 |
247868_at |
AT5G57620
|
AtMYB36, myb domain protein 36, MYB36, myb domain protein 36 |
0.206 |
0.008 |
| 71 |
266265_at |
AT2G29340
|
[NAD-dependent epimerase/dehydratase family protein] |
0.204 |
0.185 |
| 72 |
249881_at |
AT5G23190
|
CYP86B1, cytochrome P450, family 86, subfamily B, polypeptide 1 |
0.202 |
0.039 |
| 73 |
246463_at |
AT5G16970
|
AER, alkenal reductase, AT-AER, alkenal reductase |
0.202 |
0.260 |
| 74 |
265334_at |
AT2G18370
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.196 |
-0.053 |
| 75 |
256386_at |
AT1G66540
|
[Cytochrome P450 superfamily protein] |
0.196 |
-0.160 |
| 76 |
261192_at |
AT1G32870
|
ANAC013, Arabidopsis NAC domain containing protein 13, ANAC13, NAC domain protein 13, NAC13, NAC domain protein 13 |
0.195 |
-0.092 |
| 77 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.195 |
0.655 |
| 78 |
250670_at |
AT5G06860
|
ATPGIP1, POLYGALACTURONASE INHIBITING PROTEIN 1, PGIP1, polygalacturonase inhibiting protein 1 |
0.194 |
0.629 |
| 79 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.194 |
-0.016 |
| 80 |
247848_at |
AT5G58120
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.193 |
-0.045 |
| 81 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.192 |
0.361 |
| 82 |
258275_at |
AT3G15760
|
unknown |
0.191 |
0.026 |
| 83 |
264868_at |
AT1G24095
|
[Putative thiol-disulphide oxidoreductase DCC] |
0.189 |
0.105 |
| 84 |
262317_at |
AT2G48140
|
EDA4, embryo sac development arrest 4 |
0.189 |
0.026 |
| 85 |
252639_at |
AT3G44550
|
FAR5, fatty acid reductase 5 |
0.182 |
0.705 |
| 86 |
248537_at |
AT5G50100
|
[Putative thiol-disulphide oxidoreductase DCC] |
0.181 |
-0.038 |
| 87 |
262325_at |
AT1G64160
|
AtDIR5, DIR5, dirigent protein 5 |
0.181 |
0.477 |
| 88 |
247109_at |
AT5G65870
|
ATPSK5, phytosulfokine 5 precursor, PSK5, PSK5, phytosulfokine 5 precursor |
0.181 |
0.560 |
| 89 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
0.179 |
0.690 |
| 90 |
266708_at |
AT2G03200
|
[Eukaryotic aspartyl protease family protein] |
0.178 |
-0.058 |
| 91 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.178 |
1.010 |
| 92 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.178 |
0.701 |
| 93 |
254496_at |
AT4G20070
|
AAH, allantoate amidohydrolase, ATAAH, allantoate amidohydrolase |
0.177 |
-0.232 |
| 94 |
248347_at |
AT5G52250
|
EFO1, EARLY FLOWERING BY OVEREXPRESSION 1, RUP1, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 |
0.176 |
0.015 |
| 95 |
255543_at |
AT4G01870
|
[tolB protein-related] |
0.176 |
0.482 |
| 96 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
0.175 |
0.168 |
| 97 |
252364_at |
AT3G48450
|
[RPM1-interacting protein 4 (RIN4) family protein] |
0.174 |
0.833 |
| 98 |
252510_at |
AT3G46270
|
[receptor protein kinase-related] |
0.173 |
-0.003 |
| 99 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
0.171 |
-0.650 |
| 100 |
252073_at |
AT3G51750
|
unknown |
0.171 |
-0.474 |
| 101 |
259987_at |
AT1G75030
|
ATLP-3, thaumatin-like protein 3, TLP-3, thaumatin-like protein 3 |
0.171 |
-0.118 |
| 102 |
264635_at |
AT1G65500
|
unknown |
0.168 |
0.405 |
| 103 |
264124_at |
AT1G79360
|
ATOCT2, organic cation/carnitine transporter 2, OCT2, ORGANIC CATION TRANSPORTER 2, OCT2, organic cation/carnitine transporter 2 |
0.167 |
-0.029 |
| 104 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
0.166 |
0.104 |
| 105 |
261712_at |
AT1G32780
|
[GroES-like zinc-binding dehydrogenase family protein] |
0.162 |
-0.405 |
| 106 |
258227_at |
AT3G15620
|
UVR3, UV REPAIR DEFECTIVE 3 |
0.162 |
-0.052 |
| 107 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
0.160 |
-0.561 |
| 108 |
256324_at |
AT1G66760
|
[MATE efflux family protein] |
0.159 |
0.640 |
| 109 |
258452_at |
AT3G22370
|
AOX1A, alternative oxidase 1A, ATAOX1A, AtHSR3, HSR3, hyper-sensitivity-related 3 |
0.159 |
0.174 |
| 110 |
254424_at |
AT4G21510
|
AtFBS2, FBS2, F-box stress induced 2 |
0.158 |
0.111 |
| 111 |
261899_at |
AT1G80820
|
ATCCR2, CCR2, cinnamoyl coa reductase |
0.158 |
1.003 |
| 112 |
245349_at |
AT4G16690
|
ATMES16, ARABIDOPSIS THALIANA METHYL ESTERASE 16, MES16, methyl esterase 16 |
0.157 |
-0.140 |
| 113 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.153 |
0.542 |
| 114 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
0.152 |
-0.312 |
| 115 |
257919_at |
AT3G23250
|
ATMYB15, MYB DOMAIN PROTEIN 15, ATY19, MYB15, myb domain protein 15 |
0.151 |
1.110 |
| 116 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
0.150 |
0.013 |
| 117 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.150 |
0.441 |
| 118 |
255999_at |
AT1G29860
|
ATWRKY71, WRKY DNA-BINDING PROTEIN 71, WRKY71, WRKY DNA-binding protein 71 |
0.149 |
0.080 |
| 119 |
254652_at |
AT4G18170
|
ATWRKY28, WRKY28, WRKY DNA-binding protein 28 |
0.148 |
1.135 |
| 120 |
254638_at |
AT4G18740
|
[Rho termination factor] |
0.148 |
-0.339 |
| 121 |
266353_at |
AT2G01520
|
MLP328, MLP-like protein 328, ZCE1, (Zusammen-CA)-enhanced 1 |
0.147 |
-0.036 |
| 122 |
252834_at |
AT4G40070
|
[RING/U-box superfamily protein] |
0.147 |
0.083 |
| 123 |
261509_at |
AT1G71740
|
unknown |
0.147 |
-0.053 |
| 124 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
0.145 |
0.558 |
| 125 |
248160_at |
AT5G54470
|
BBX29, B-box domain protein 29 |
0.145 |
0.416 |
| 126 |
247731_at |
AT5G59590
|
UGT76E2, UDP-glucosyl transferase 76E2 |
0.144 |
0.127 |
| 127 |
248952_at |
AT5G45410
|
unknown |
0.144 |
0.095 |
| 128 |
259970_at |
AT1G76570
|
AtLHCB7, LHCB7, light-harvesting complex B7 |
0.143 |
-0.109 |
| 129 |
259705_at |
AT1G77450
|
anac032, NAC domain containing protein 32, NAC032, NAC domain containing protein 32 |
0.143 |
0.512 |
| 130 |
261618_at |
AT1G33110
|
[MATE efflux family protein] |
0.141 |
0.025 |
| 131 |
267083_at |
AT2G41100
|
ATCAL4, ARABIDOPSIS THALIANA CALMODULIN LIKE 4, TCH3, TOUCH 3 |
0.141 |
0.224 |
| 132 |
249576_at |
AT5G37690
|
[SGNH hydrolase-type esterase superfamily protein] |
0.141 |
-0.033 |
| 133 |
246396_at |
AT1G58180
|
ATBCA6, A. THALIANA BETA CARBONIC ANHYDRASE 6, BCA6, beta carbonic anhydrase 6 |
0.139 |
0.082 |
| 134 |
247035_at |
AT5G67110
|
ALC, ALCATRAZ |
0.139 |
-0.023 |
| 135 |
246310_at |
AT3G51895
|
AST12, SULTR3;1, sulfate transporter 3;1 |
0.139 |
-0.058 |
| 136 |
262635_at |
AT1G06570
|
HPD, 4-hydroxyphenylpyruvate dioxygenase, HPPD, 4-hydroxyphenylpyruvate dioxygenase, PDS1, phytoene desaturation 1 |
0.138 |
-0.021 |
| 137 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.138 |
0.347 |
| 138 |
260205_at |
AT1G70700
|
JAZ9, JASMONATE-ZIM-DOMAIN PROTEIN 9, TIFY7 |
0.138 |
0.397 |
| 139 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
0.137 |
-0.152 |
| 140 |
248199_at |
AT5G54170
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.137 |
0.418 |
| 141 |
246034_at |
AT5G08350
|
[GRAM domain-containing protein / ABA-responsive protein-related] |
0.136 |
-0.256 |
| 142 |
258315_at |
AT3G16175
|
[Thioesterase superfamily protein] |
0.136 |
-0.237 |
| 143 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.136 |
0.844 |
| 144 |
251774_at |
AT3G55840
|
[Hs1pro-1 protein] |
0.133 |
0.559 |
| 145 |
264010_at |
AT2G21100
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.133 |
0.014 |
| 146 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
0.132 |
1.244 |
| 147 |
248207_at |
AT5G53970
|
TAT7, tyrosine aminotransferase 7 |
0.132 |
-0.068 |
| 148 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
0.132 |
0.405 |
| 149 |
267181_at |
AT2G37760
|
AKR4C8, Aldo-keto reductase family 4 member C8 |
0.131 |
0.178 |
| 150 |
262916_at |
AT1G59700
|
ATGSTU16, glutathione S-transferase TAU 16, GSTU16, glutathione S-transferase TAU 16 |
0.131 |
0.039 |
| 151 |
245713_at |
AT5G04370
|
NAMT1 |
0.130 |
0.105 |
| 152 |
249406_at |
AT5G40210
|
UMAMIT42, Usually multiple acids move in and out Transporters 42 |
0.130 |
0.354 |
| 153 |
267364_at |
AT2G40080
|
ELF4, EARLY FLOWERING 4 |
0.129 |
0.172 |
| 154 |
256096_at |
AT1G13650
|
unknown |
-0.526 |
-0.608 |
| 155 |
250561_at |
AT5G08030
|
AtGDPD6, GDPD6, glycerophosphodiester phosphodiesterase 6 |
-0.488 |
-0.014 |
| 156 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
-0.425 |
-0.404 |
| 157 |
246652_at |
AT5G35190
|
EXT13, extensin 13 |
-0.366 |
-0.058 |
| 158 |
265414_at |
AT2G16660
|
[Major facilitator superfamily protein] |
-0.338 |
0.189 |
| 159 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
-0.334 |
0.487 |
| 160 |
263715_at |
AT2G20570
|
GBF's pro-rich region-interacting factor 1 |
-0.333 |
-0.075 |
| 161 |
257793_at |
AT3G26960
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.296 |
-0.564 |
| 162 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
-0.291 |
0.749 |
| 163 |
251226_at |
AT3G62680
|
ATPRP3, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 3, PRP3, proline-rich protein 3 |
-0.289 |
0.047 |
| 164 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.287 |
0.048 |
| 165 |
255284_at |
AT4G04610
|
APR, APR1, APS reductase 1, ATAPR1, PRH19, PAPS REDUCTASE HOMOLOG 19 |
-0.284 |
-0.124 |
| 166 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
-0.273 |
0.029 |
| 167 |
246991_at |
AT5G67400
|
RHS19, root hair specific 19 |
-0.268 |
-0.057 |
| 168 |
252437_at |
AT3G47380
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.259 |
0.643 |
| 169 |
264341_at |
AT1G70270
|
unknown |
-0.257 |
-0.267 |
| 170 |
254931_at |
AT4G11460
|
CRK30, cysteine-rich RLK (RECEPTOR-like protein kinase) 30 |
-0.253 |
-0.270 |
| 171 |
257201_at |
AT3G23740
|
unknown |
-0.252 |
-0.147 |
| 172 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.250 |
-0.365 |
| 173 |
259481_at |
AT1G18970
|
GLP4, germin-like protein 4 |
-0.245 |
-0.007 |
| 174 |
251476_at |
AT3G59670
|
unknown |
-0.244 |
-0.212 |
| 175 |
248277_at |
AT5G52860
|
ABCG8, ATP-binding cassette G8 |
-0.240 |
0.014 |
| 176 |
264114_at |
AT2G31270
|
ATCDT1A, ARABIDOPSIS HOMOLOG OF YEAST CDT1 A, CDT1, ARABIDOPSIS HOMOLOG OF YEAST CDT1, CDT1A, homolog of yeast CDT1 A |
-0.234 |
-0.100 |
| 177 |
253788_at |
AT4G28680
|
AtTYDC, TYDC, L-tyrosine decarboxylase, TYRDC, L-tyrosine decarboxylase, TYRDC1, L-TYROSINE DECARBOXYLASE 1 |
-0.232 |
0.050 |
| 178 |
248961_at |
AT5G45650
|
[subtilase family protein] |
-0.232 |
-0.398 |
| 179 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
-0.230 |
-0.017 |
| 180 |
248540_at |
AT5G50160
|
ATFRO8, FERRIC REDUCTION OXIDASE 8, FRO8, ferric reduction oxidase 8 |
-0.228 |
0.042 |
| 181 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
-0.226 |
0.005 |
| 182 |
246241_at |
AT4G37050
|
AtPLAIVC, phospholipase A IVC, PLA V, PLAIII{beta}, patatin-related phospholipase III beta, PLP4, PATATIN-like protein 4 |
-0.224 |
-0.094 |
| 183 |
259656_at |
AT1G55200
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.221 |
-0.060 |
| 184 |
266465_at |
AT2G47750
|
GH3.9, putative indole-3-acetic acid-amido synthetase GH3.9 |
-0.220 |
-0.129 |
| 185 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.220 |
-0.047 |
| 186 |
252916_at |
AT4G38950
|
[ATP binding microtubule motor family protein] |
-0.216 |
-0.588 |
| 187 |
251952_at |
AT3G53650
|
[Histone superfamily protein] |
-0.215 |
-0.018 |
| 188 |
251287_at |
AT3G61820
|
[Eukaryotic aspartyl protease family protein] |
-0.211 |
-0.003 |
| 189 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
-0.205 |
0.065 |
| 190 |
254235_at |
AT4G23750
|
CRF2, cytokinin response factor 2, TMO3, TARGET OF MONOPTEROS 3 |
-0.195 |
-0.290 |
| 191 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.194 |
0.312 |
| 192 |
252361_at |
AT3G48490
|
unknown |
-0.193 |
-0.078 |
| 193 |
247056_at |
AT5G66750
|
ATDDM1, CHA1, CHR01, CHROMATIN REMODELING 1, CHR1, chromatin remodeling 1, DDM1, DECREASED DNA METHYLATION 1, SOM1, SOMNIFEROUS 1, SOM4 |
-0.193 |
-0.207 |
| 194 |
249340_at |
AT5G41140
|
[Myosin heavy chain-related protein] |
-0.193 |
-0.680 |
| 195 |
253514_at |
AT4G31805
|
POLAR, POLAR LOCALIZATION DURING ASYMMETRIC DIVISION AND REDISTRIBUTION |
-0.190 |
-0.010 |
| 196 |
257460_at |
AT1G75580
|
SAUR51, SMALL AUXIN UPREGULATED RNA 51 |
-0.190 |
0.418 |
| 197 |
260717_at |
AT1G48120
|
[hydrolases] |
-0.190 |
-0.467 |
| 198 |
265097_at |
AT1G04020
|
ATBARD1, BARD1, breast cancer associated RING 1, ROW1 |
-0.190 |
-0.208 |
| 199 |
250560_at |
AT5G08020
|
ATRPA70B, ARABIDOPSIS THALIANA RPA70-KDA SUBUNIT B, RPA70B, RPA70-kDa subunit B |
-0.188 |
-0.088 |
| 200 |
261330_at |
AT1G44900
|
ATMCM2, MCM2, MINICHROMOSOME MAINTENANCE 2 |
-0.188 |
-0.291 |
| 201 |
247549_at |
AT5G61420
|
AtMYB28, HAG1, HIGH ALIPHATIC GLUCOSINOLATE 1, MYB28, myb domain protein 28, PMG1, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 1 |
-0.187 |
-0.411 |
| 202 |
259653_at |
AT1G55240
|
unknown |
-0.187 |
0.015 |
| 203 |
247606_at |
AT5G61000
|
ATRPA70D, RPA70D |
-0.187 |
-0.170 |
| 204 |
258895_at |
AT3G05600
|
[alpha/beta-Hydrolases superfamily protein] |
-0.184 |
0.057 |
| 205 |
266912_at |
AT2G45900
|
TRM13, TON1 Recruiting Motif 13 |
-0.183 |
-0.271 |
| 206 |
255856_at |
AT1G66940
|
[protein kinase-related] |
-0.183 |
-0.227 |
| 207 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
-0.183 |
0.293 |
| 208 |
255450_at |
AT4G02850
|
[phenazine biosynthesis PhzC/PhzF family protein] |
-0.182 |
-0.204 |
| 209 |
257571_at |
AT3G16870
|
GATA17, GATA transcription factor 17 |
-0.181 |
0.136 |
| 210 |
251593_at |
AT3G57660
|
NRPA1, nuclear RNA polymerase A1 |
-0.181 |
-0.176 |
| 211 |
260192_at |
AT1G67630
|
EMB2814, EMBRYO DEFECTIVE 2814, POLA2, DNA polymerase alpha 2 |
-0.179 |
-0.213 |
| 212 |
249347_at |
AT5G40830
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.177 |
-0.271 |
| 213 |
252911_at |
AT4G39510
|
CYP96A12, cytochrome P450, family 96, subfamily A, polypeptide 12 |
-0.176 |
-0.513 |
| 214 |
254490_at |
AT4G20320
|
[CTP synthase family protein] |
-0.174 |
0.519 |
| 215 |
266089_at |
AT2G38010
|
[Neutral/alkaline non-lysosomal ceramidase] |
-0.173 |
0.088 |
| 216 |
248841_at |
AT5G46740
|
UBP21, ubiquitin-specific protease 21 |
-0.171 |
-0.071 |
| 217 |
267173_at |
AT2G37560
|
ATORC2, ORIGIN RECOGNITION COMPLEX SECOND LARGEST SUBUNIT 2, ORC2, origin recognition complex second largest subunit 2 |
-0.171 |
-0.042 |
| 218 |
261768_at |
AT1G15550
|
ATGA3OX1, ARABIDOPSIS THALIANA GIBBERELLIN 3 BETA-HYDROXYLASE 1, GA3OX1, gibberellin 3-oxidase 1, GA4, GA REQUIRING 4 |
-0.168 |
-0.257 |
| 219 |
247168_at |
AT5G65860
|
[ankyrin repeat family protein] |
-0.168 |
-0.002 |
| 220 |
247871_at |
AT5G57530
|
AtXTH12, XTH12, xyloglucan endotransglucosylase/hydrolase 12 |
-0.167 |
0.031 |
| 221 |
251638_at |
AT3G57490
|
[Ribosomal protein S5 family protein] |
-0.166 |
0.073 |
| 222 |
259303_at |
AT3G05130
|
unknown |
-0.165 |
0.139 |
| 223 |
250762_at |
AT5G05990
|
[Mitochondrial glycoprotein family protein] |
-0.165 |
-0.117 |
| 224 |
246983_at |
AT5G67200
|
[Leucine-rich repeat protein kinase family protein] |
-0.163 |
-0.301 |
| 225 |
262543_at |
AT1G34245
|
EPF2, EPIDERMAL PATTERNING FACTOR 2 |
-0.163 |
-0.120 |
| 226 |
265730_at |
AT2G32220
|
[Ribosomal L27e protein family] |
-0.163 |
0.098 |
| 227 |
257115_at |
AT3G20150
|
[Kinesin motor family protein] |
-0.162 |
-0.068 |
| 228 |
249788_at |
AT5G24330
|
ATXR6, ARABIDOPSIS TRITHORAX-RELATED PROTEIN 6, SDG34, SET DOMAIN PROTEIN 34 |
-0.160 |
-0.044 |
| 229 |
256675_at |
AT3G52170
|
[DNA binding] |
-0.157 |
-0.188 |
| 230 |
254379_at |
AT4G21820
|
[binding] |
-0.157 |
-0.061 |
| 231 |
259632_at |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
-0.156 |
-0.524 |
| 232 |
250327_at |
AT5G12050
|
unknown |
-0.155 |
-0.252 |
| 233 |
255403_at |
AT4G03400
|
DFL2, DWARF IN LIGHT 2, GH3-10 |
-0.154 |
0.464 |
| 234 |
255410_at |
AT4G03100
|
[Rho GTPase activating protein with PAK-box/P21-Rho-binding domain] |
-0.153 |
-0.147 |
| 235 |
266640_at |
AT2G35585
|
unknown |
-0.153 |
-0.063 |
| 236 |
264969_at |
AT1G67320
|
EMB2813, EMBRYO DEFECTIVE 2813 |
-0.153 |
-0.115 |
| 237 |
251982_at |
AT3G53190
|
[Pectin lyase-like superfamily protein] |
-0.153 |
-0.212 |
| 238 |
254043_at |
AT4G25990
|
CIL |
-0.153 |
-0.282 |
| 239 |
258024_at |
AT3G19360
|
[Zinc finger (CCCH-type) family protein] |
-0.152 |
0.028 |
| 240 |
265695_at |
AT2G24490
|
ATRPA2, ATRPA32A, ROR1, SUPPRESSOR OF ROS1, RPA2, replicon protein A2, RPA32A |
-0.152 |
-0.051 |
| 241 |
246898_at |
AT5G25580
|
unknown |
-0.151 |
-0.090 |
| 242 |
262752_at |
AT1G16330
|
CYCB3;1, cyclin b3;1 |
-0.150 |
-0.026 |
| 243 |
267012_at |
AT2G39220
|
PLA IIB, PLP6, PATATIN-like protein 6 |
-0.150 |
-0.075 |
| 244 |
249129_at |
AT5G43080
|
CYCA3;1, Cyclin A3;1 |
-0.149 |
-0.112 |
| 245 |
263034_at |
AT1G24020
|
MLP423, MLP-like protein 423 |
-0.149 |
-0.111 |
| 246 |
258898_at |
AT3G05740
|
RECQI1, RECQ helicase l1 |
-0.148 |
-0.077 |
| 247 |
252468_at |
AT3G46970
|
ATPHS2, Arabidopsis thaliana alpha-glucan phosphorylase 2, PHS2, alpha-glucan phosphorylase 2 |
-0.147 |
0.084 |
| 248 |
256746_at |
AT3G29320
|
PHS1, alpha-glucan phosphorylase 1 |
-0.147 |
0.113 |
| 249 |
254065_at |
AT4G25420
|
AT2301, ATGA20OX1, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 1, GA20OX1, GA5, GA REQUIRING 5 |
-0.146 |
-0.500 |
| 250 |
248888_at |
AT5G46240
|
AtKAT1, KAT1, potassium channel in Arabidopsis thaliana 1 |
-0.146 |
-0.002 |
| 251 |
248139_at |
AT5G54970
|
unknown |
-0.145 |
0.234 |
| 252 |
250379_at |
AT5G11590
|
TINY2, TINY2 |
-0.145 |
-0.284 |
| 253 |
248372_at |
AT5G51850
|
TRM24, TON1 Recruiting Motif 24 |
-0.145 |
-0.277 |
| 254 |
260417_at |
AT1G69770
|
CMT3, chromomethylase 3 |
-0.145 |
-0.085 |
| 255 |
256721_at |
AT2G34150
|
ATRANGAP2, ATSCAR1, SCAR1, WAVE1, WISKOTT-ALDRICH SYNDROME PROTEIN FAMILY VERPROLIN HOMOLOGOUS PROTEIN 1 |
-0.144 |
-0.446 |
| 256 |
248891_at |
AT5G46280
|
MCM3, MINICHROMOSOME MAINTENANCE 3 |
-0.143 |
-0.269 |
| 257 |
252442_at |
AT3G46940
|
DUT1, DUTP-PYROPHOSPHATASE-LIKE 1 |
-0.143 |
-0.122 |
| 258 |
266510_at |
AT2G47990
|
EDA13, EMBRYO SAC DEVELOPMENT ARREST 13, EDA19, EMBRYO SAC DEVELOPMENT ARREST 19, SWA1, SLOW WALKER1 |
-0.141 |
-0.017 |
| 259 |
247739_at |
AT5G59240
|
[Ribosomal protein S8e family protein] |
-0.140 |
0.028 |
| 260 |
245818_at |
AT1G26100
|
[Cytochrome b561/ferric reductase transmembrane protein family] |
-0.140 |
-0.177 |
| 261 |
245133_at |
AT2G45310
|
GAE4, UDP-D-glucuronate 4-epimerase 4 |
-0.140 |
-0.185 |
| 262 |
261080_at |
AT1G07370
|
ATPCNA1, PROLIFERATING CELLULAR NUCLEAR ANTIGEN 1, PCNA1, proliferating cellular nuclear antigen 1 |
-0.140 |
-0.149 |
| 263 |
255648_at |
AT4G00910
|
[Aluminium activated malate transporter family protein] |
-0.138 |
-0.019 |
| 264 |
258529_at |
AT3G06740
|
GATA15, GATA transcription factor 15 |
-0.138 |
-0.258 |
| 265 |
262766_at |
AT1G13160
|
[ARM repeat superfamily protein] |
-0.137 |
-0.061 |
| 266 |
257663_at |
AT3G20260
|
[FUNCTIONS IN: structural constituent of ribosome] |
-0.137 |
-0.230 |
| 267 |
263913_at |
AT2G36570
|
PXC1, PXY/TDR-correlated 1 |
-0.137 |
-0.432 |
| 268 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
-0.137 |
0.602 |
| 269 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.137 |
-0.621 |
| 270 |
255645_at |
AT4G00880
|
SAUR31, SMALL AUXIN UPREGULATED RNA 31 |
-0.137 |
-0.064 |
| 271 |
258138_at |
AT3G24495
|
ATMSH7, ARABIDOPSIS THALIANA MUTS HOMOLOG 7, MSH6-2, MUTS HOMOLOG 6-2, MSH7, MUTS homolog 7 |
-0.135 |
-0.205 |
| 272 |
248614_at |
AT5G49560
|
[Putative methyltransferase family protein] |
-0.135 |
-0.168 |
| 273 |
265311_at |
AT2G20250
|
unknown |
-0.135 |
-0.049 |
| 274 |
262758_at |
AT1G10780
|
[F-box/RNI-like superfamily protein] |
-0.134 |
-0.105 |
| 275 |
246315_at |
AT3G56870
|
unknown |
-0.134 |
-0.045 |
| 276 |
267497_at |
AT2G30540
|
[Thioredoxin superfamily protein] |
-0.134 |
-0.049 |
| 277 |
259278_at |
AT3G01160
|
unknown |
-0.133 |
-0.023 |
| 278 |
255887_at |
AT1G20370
|
[Pseudouridine synthase family protein] |
-0.132 |
-0.072 |
| 279 |
267382_at |
AT2G44300
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.132 |
-0.185 |
| 280 |
251368_at |
AT3G61380
|
TRM14, TON1 Recruiting Motif 14 |
-0.132 |
-0.163 |
| 281 |
255513_at |
AT4G02060
|
MCM7, PRL, PROLIFERA |
-0.131 |
-0.208 |
| 282 |
260276_at |
AT1G80450
|
[VQ motif-containing protein] |
-0.131 |
0.697 |
| 283 |
263612_at |
AT2G16440
|
MCM4, MINICHROMOSOME MAINTENANCE 4 |
-0.131 |
-0.284 |
| 284 |
264783_at |
AT1G08650
|
ATPPCK1, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 1, PPCK1, phosphoenolpyruvate carboxylase kinase 1 |
-0.131 |
-0.013 |
| 285 |
262313_at |
AT1G70900
|
unknown |
-0.131 |
-0.152 |
| 286 |
264899_at |
AT1G23130
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.131 |
-0.022 |
| 287 |
262594_at |
AT1G15250
|
[Zinc-binding ribosomal protein family protein] |
-0.130 |
-0.049 |
| 288 |
263552_x_at |
AT2G24980
|
EXT6, extensin 6 |
-0.129 |
-0.098 |
| 289 |
249080_at |
AT5G43990
|
SDG18, SET DOMAIN PROTEIN 18, SUVR2 |
-0.129 |
-0.323 |
| 290 |
248104_at |
AT5G55250
|
AtIAMT1, IAMT1, IAA carboxylmethyltransferase 1 |
-0.129 |
-0.346 |
| 291 |
246986_at |
AT5G67280
|
RLK, receptor-like kinase |
-0.128 |
-0.292 |
| 292 |
255874_at |
AT2G40550
|
ETG1, E2F target gene 1 |
-0.127 |
-0.143 |
| 293 |
251620_at |
AT3G58060
|
[Cation efflux family protein] |
-0.127 |
-0.081 |
| 294 |
253454_at |
AT4G31875
|
unknown |
-0.127 |
-0.148 |
| 295 |
245411_at |
AT4G17240
|
unknown |
-0.126 |
0.299 |
| 296 |
262002_at |
AT1G64450
|
[Glycine-rich protein family] |
-0.126 |
-0.170 |
| 297 |
257294_at |
AT3G15570
|
[Phototropic-responsive NPH3 family protein] |
-0.126 |
-0.361 |
| 298 |
261804_at |
AT1G30530
|
UGT78D1, UDP-glucosyl transferase 78D1 |
-0.126 |
-0.235 |
| 299 |
249134_at |
AT5G43150
|
unknown |
-0.125 |
-0.112 |
| 300 |
245828_at |
AT1G57820
|
ORTH2, ORTHRUS 2, VIM1, VARIANT IN METHYLATION 1 |
-0.125 |
-0.052 |
| 301 |
253817_at |
AT4G28310
|
unknown |
-0.125 |
-0.127 |
| 302 |
252400_at |
AT3G48020
|
unknown |
-0.125 |
0.463 |
| 303 |
254371_at |
AT4G21760
|
BGLU47, beta-glucosidase 47 |
-0.124 |
-0.612 |
| 304 |
250045_at |
AT5G17700
|
[MATE efflux family protein] |
-0.124 |
-0.179 |
| 305 |
264818_at |
AT1G03530
|
ATNAF1, NAF1, nuclear assembly factor 1 |
-0.124 |
-0.181 |