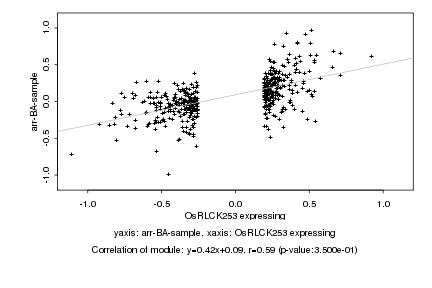
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
OsRLCK253 expressing |
Log2 signal ratio
arr-BA-sample |
| 1 |
257057_at |
AT3G15310
|
unknown |
0.920 |
0.616 |
| 2 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
0.710 |
0.657 |
| 3 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
0.708 |
0.357 |
| 4 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
0.659 |
0.685 |
| 5 |
260380_at |
AT1G73870
|
BBX16, B-box domain protein 16, COL7, CONSTANS-LIKE 7 |
0.651 |
0.471 |
| 6 |
264846_at |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
0.570 |
0.323 |
| 7 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
0.547 |
0.632 |
| 8 |
265698_at |
AT2G32160
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.536 |
-0.266 |
| 9 |
260462_at |
AT1G10970
|
ATZIP4, ZIP4, zinc transporter 4 precursor |
0.534 |
0.539 |
| 10 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
0.532 |
0.147 |
| 11 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
0.529 |
0.565 |
| 12 |
256096_at |
AT1G13650
|
unknown |
0.521 |
0.069 |
| 13 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
0.515 |
0.969 |
| 14 |
254574_at |
AT4G19430
|
unknown |
0.509 |
0.096 |
| 15 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
0.507 |
0.796 |
| 16 |
265939_at |
AT2G19650
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.502 |
0.639 |
| 17 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
0.501 |
0.421 |
| 18 |
256779_at |
AT3G13784
|
AtcwINV5, cell wall invertase 5, CWINV5, cell wall invertase 5 |
0.499 |
0.162 |
| 19 |
253496_at |
AT4G31870
|
ATGPX7, GLUTATHIONE PEROXIDASE 7, GPX7, glutathione peroxidase 7 |
0.485 |
-0.233 |
| 20 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
0.482 |
0.137 |
| 21 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
0.471 |
0.922 |
| 22 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
0.466 |
0.382 |
| 23 |
252661_at |
AT3G44450
|
unknown |
0.458 |
0.276 |
| 24 |
260419_at |
AT1G69730
|
[Wall-associated kinase family protein] |
0.456 |
0.253 |
| 25 |
261150_at |
AT1G19640
|
JMT, jasmonic acid carboxyl methyltransferase |
0.451 |
-0.125 |
| 26 |
247055_at |
AT5G66740
|
unknown |
0.451 |
0.183 |
| 27 |
265892_at |
AT2G15020
|
unknown |
0.437 |
0.557 |
| 28 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
0.430 |
0.616 |
| 29 |
251869_at |
AT3G54500
|
LNK2, night light-inducible and clock-regulated 2 |
0.429 |
0.408 |
| 30 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
0.420 |
0.799 |
| 31 |
264261_at |
AT1G09240
|
ATNAS3, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 3, NAS3, nicotianamine synthase 3 |
0.418 |
0.130 |
| 32 |
245982_at |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
0.415 |
0.808 |
| 33 |
250665_at |
AT5G06980
|
LNK4, night light-inducible and clock-regulated 4 |
0.412 |
0.528 |
| 34 |
259927_at |
AT1G75100
|
JAC1, J-domain protein required for chloroplast accumulation response 1 |
0.411 |
0.447 |
| 35 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
0.407 |
0.448 |
| 36 |
264436_at |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
0.405 |
0.226 |
| 37 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
0.404 |
0.587 |
| 38 |
254145_at |
AT4G24700
|
unknown |
0.402 |
0.405 |
| 39 |
263739_at |
AT2G21320
|
BBX18, B-box domain protein 18 |
0.397 |
0.499 |
| 40 |
264958_at |
AT1G76960
|
unknown |
0.394 |
-0.100 |
| 41 |
252520_at |
AT3G46370
|
[Leucine-rich repeat protein kinase family protein] |
0.385 |
-0.051 |
| 42 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
0.384 |
0.151 |
| 43 |
259042_at |
AT3G03450
|
RGL2, RGA-like 2 |
0.383 |
0.113 |
| 44 |
263796_at |
AT2G24540
|
AFR, ATTENUATED FAR-RED RESPONSE |
0.379 |
0.259 |
| 45 |
252414_at |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
0.378 |
0.291 |
| 46 |
248246_at |
AT5G53200
|
TRY, TRIPTYCHON |
0.369 |
-0.006 |
| 47 |
262826_at |
AT1G13080
|
CYP71B2, cytochrome P450, family 71, subfamily B, polypeptide 2 |
0.368 |
0.015 |
| 48 |
256924_at |
AT3G29590
|
AT5MAT |
0.365 |
0.649 |
| 49 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
0.363 |
-0.008 |
| 50 |
261407_at |
AT1G18810
|
[phytochrome kinase substrate-related] |
0.362 |
0.176 |
| 51 |
259723_at |
AT1G60960
|
ATIRT3, IRON REGULATED TRANSPORTER 3, IRT3, iron regulated transporter 3 |
0.362 |
0.230 |
| 52 |
250344_at |
AT5G11930
|
[Thioredoxin superfamily protein] |
0.361 |
0.371 |
| 53 |
261958_at |
AT1G64500
|
[Glutaredoxin family protein] |
0.358 |
0.544 |
| 54 |
263252_at |
AT2G31380
|
BBX25, B-box domain protein 25, STH, salt tolerance homologue |
0.352 |
0.580 |
| 55 |
247452_at |
AT5G62430
|
CDF1, cycling DOF factor 1 |
0.352 |
0.336 |
| 56 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.342 |
0.931 |
| 57 |
259560_at |
AT1G21270
|
WAK2, wall-associated kinase 2 |
0.338 |
0.367 |
| 58 |
247602_at |
AT5G60900
|
RLK1, receptor-like protein kinase 1 |
0.334 |
-0.095 |
| 59 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
0.330 |
0.294 |
| 60 |
245152_at |
AT2G47490
|
ATNDT1, ARABIDOPSIS THALIANA NAD+ TRANSPORTER 1, NDT1, NAD+ transporter 1 |
0.326 |
0.212 |
| 61 |
253298_at |
AT4G33560
|
[Wound-responsive family protein] |
0.326 |
0.484 |
| 62 |
261339_at |
AT1G35710
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.325 |
0.299 |
| 63 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.323 |
-0.352 |
| 64 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
0.322 |
0.760 |
| 65 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
0.319 |
-0.079 |
| 66 |
252916_at |
AT4G38950
|
[ATP binding microtubule motor family protein] |
0.318 |
-0.100 |
| 67 |
255503_at |
AT4G02420
|
LecRK-IV.4, L-type lectin receptor kinase IV.4 |
0.315 |
0.227 |
| 68 |
247463_at |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
0.313 |
-0.125 |
| 69 |
246523_at |
AT5G15850
|
ATCOL1, BBX2, B-box domain protein 2, COL1, CONSTANS-like 1 |
0.313 |
0.368 |
| 70 |
246597_at |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
0.313 |
0.493 |
| 71 |
249798_at |
AT5G23730
|
EFO2, EARLY FLOWERING BY OVEREXPRESSION 2, RUP2, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 2 |
0.312 |
0.080 |
| 72 |
259275_at |
AT3G01060
|
unknown |
0.308 |
0.290 |
| 73 |
249675_at |
AT5G35940
|
[Mannose-binding lectin superfamily protein] |
0.306 |
-0.145 |
| 74 |
265122_at |
AT1G62540
|
FMO GS-OX2, flavin-monooxygenase glucosinolate S-oxygenase 2 |
0.302 |
0.201 |
| 75 |
260603_at |
AT1G55960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.300 |
0.188 |
| 76 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
0.297 |
0.083 |
| 77 |
250558_at |
AT5G07990
|
CYP75B1, CYTOCHROME P450 75B1, D501, TT7, TRANSPARENT TESTA 7 |
0.297 |
0.421 |
| 78 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
0.293 |
0.304 |
| 79 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.292 |
0.332 |
| 80 |
254371_at |
AT4G21760
|
BGLU47, beta-glucosidase 47 |
0.292 |
-0.241 |
| 81 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
0.290 |
0.383 |
| 82 |
254250_at |
AT4G23290
|
CRK21, cysteine-rich RLK (RECEPTOR-like protein kinase) 21 |
0.288 |
0.224 |
| 83 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
0.288 |
0.034 |
| 84 |
248961_at |
AT5G45650
|
[subtilase family protein] |
0.287 |
0.102 |
| 85 |
255694_at |
AT4G00050
|
UNE10, unfertilized embryo sac 10 |
0.287 |
0.280 |
| 86 |
263777_at |
AT2G46450
|
ATCNGC12, cyclic nucleotide-gated channel 12, CNGC12, cyclic nucleotide-gated channel 12 |
0.282 |
0.321 |
| 87 |
261068_at |
AT1G07450
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.282 |
0.225 |
| 88 |
248160_at |
AT5G54470
|
BBX29, B-box domain protein 29 |
0.281 |
0.121 |
| 89 |
252746_at |
AT3G43190
|
ATSUS4, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 4, SUS4, sucrose synthase 4 |
0.281 |
0.186 |
| 90 |
266738_at |
AT2G47010
|
unknown |
0.280 |
0.317 |
| 91 |
255763_at |
AT1G16730
|
unknown |
0.279 |
0.310 |
| 92 |
267282_at |
AT2G19390
|
unknown |
0.279 |
0.086 |
| 93 |
247232_at |
AT5G64940
|
ATATH13, ARABIDOPSIS THALIANA ABC2 HOMOLOG 13, ATH13, ABC2 homolog 13, ATOSA1, A. THALIANA OXIDATIVE STRESS-RELATED ABC1-LIKE PROTEIN 1, OSA1, OXIDATIVE STRESS-RELATED ABC1-LIKE PROTEIN 1 |
0.279 |
0.340 |
| 94 |
249677_at |
AT5G35970
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.278 |
0.185 |
| 95 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
0.275 |
0.413 |
| 96 |
248333_at |
AT5G52390
|
[PAR1 protein] |
0.272 |
0.148 |
| 97 |
254746_at |
AT4G12980
|
[Auxin-responsive family protein] |
0.271 |
0.090 |
| 98 |
245242_at |
AT1G44446
|
ATCAO, ARABIDOPSIS THALIANA CHLOROPHYLL A OXYGENASE, CAO, CHLOROPHYLL A OXYGENASE, CH1, CHLORINA 1 |
0.270 |
0.389 |
| 99 |
257153_at |
AT3G27220
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.270 |
0.222 |
| 100 |
254185_at |
AT4G23990
|
ATCSLG3, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE-LIKE G3, CSLG3, cellulose synthase like G3 |
0.269 |
0.307 |
| 101 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
0.269 |
0.251 |
| 102 |
252619_at |
AT3G45210
|
unknown |
0.268 |
0.268 |
| 103 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
0.268 |
0.042 |
| 104 |
266336_at |
AT2G32270
|
ZIP3, zinc transporter 3 precursor |
0.268 |
-0.192 |
| 105 |
249754_at |
AT5G24530
|
DMR6, DOWNY MILDEW RESISTANT 6 |
0.266 |
0.134 |
| 106 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
0.262 |
0.316 |
| 107 |
262290_at |
AT1G70985
|
[hydroxyproline-rich glycoprotein family protein] |
0.262 |
0.788 |
| 108 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
0.262 |
0.268 |
| 109 |
247323_at |
AT5G64170
|
LNK1, night light-inducible and clock-regulated 1 |
0.262 |
0.334 |
| 110 |
266363_at |
AT2G41250
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.261 |
0.263 |
| 111 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
0.259 |
0.538 |
| 112 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
0.258 |
0.415 |
| 113 |
247214_at |
AT5G64850
|
unknown |
0.257 |
0.337 |
| 114 |
252429_at |
AT3G47500
|
CDF3, cycling DOF factor 3 |
0.257 |
0.433 |
| 115 |
266778_at |
AT2G29090
|
CYP707A2, cytochrome P450, family 707, subfamily A, polypeptide 2 |
0.257 |
-0.032 |
| 116 |
262103_at |
AT1G02940
|
ATGSTF5, GLUTATHIONE S-TRANSFERASE (CLASS PHI) 5, GSTF5, glutathione S-transferase (class phi) 5 |
0.255 |
0.258 |
| 117 |
251826_at |
AT3G55110
|
ABCG18, ATP-binding cassette G18 |
0.255 |
0.027 |
| 118 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.255 |
0.086 |
| 119 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
0.255 |
-0.180 |
| 120 |
245463_at |
AT4G17030
|
AT-EXPR, ATEXLB1, expansin-like B1, ATEXPR1, ATHEXP BETA 3.1, EXLB1, expansin-like B1, EXPR |
0.253 |
0.457 |
| 121 |
249415_at |
AT5G39660
|
CDF2, cycling DOF factor 2 |
0.253 |
0.240 |
| 122 |
253788_at |
AT4G28680
|
AtTYDC, TYDC, L-tyrosine decarboxylase, TYRDC, L-tyrosine decarboxylase, TYRDC1, L-TYROSINE DECARBOXYLASE 1 |
0.252 |
0.124 |
| 123 |
254251_at |
AT4G23300
|
CRK22, cysteine-rich RLK (RECEPTOR-like protein kinase) 22 |
0.252 |
0.424 |
| 124 |
262811_at |
AT1G11700
|
unknown |
0.252 |
0.540 |
| 125 |
259432_at |
AT1G01520
|
ASG4, ALTERED SEED GERMINATION 4 |
0.250 |
0.304 |
| 126 |
249384_at |
AT5G39890
|
unknown |
0.249 |
0.233 |
| 127 |
248037_at |
AT5G55930
|
ATOPT1, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 1, OPT1, oligopeptide transporter 1 |
0.246 |
0.373 |
| 128 |
262507_at |
AT1G11330
|
[S-locus lectin protein kinase family protein] |
0.246 |
0.066 |
| 129 |
247977_at |
AT5G56850
|
unknown |
0.245 |
-0.012 |
| 130 |
255842_at |
AT2G33530
|
scpl46, serine carboxypeptidase-like 46 |
0.245 |
0.120 |
| 131 |
256049_at |
AT1G07010
|
AtSLP1, SLP1, Shewenella-like protein phosphatase 1 |
0.244 |
0.368 |
| 132 |
261804_at |
AT1G30530
|
UGT78D1, UDP-glucosyl transferase 78D1 |
0.242 |
0.074 |
| 133 |
259632_at |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
0.242 |
0.309 |
| 134 |
253411_at |
AT4G32980
|
ATH1, homeobox gene 1 |
0.241 |
0.212 |
| 135 |
252073_at |
AT3G51750
|
unknown |
0.240 |
0.147 |
| 136 |
245558_at |
AT4G15430
|
[ERD (early-responsive to dehydration stress) family protein] |
0.240 |
0.467 |
| 137 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
0.239 |
0.096 |
| 138 |
249035_at |
AT5G44190
|
ATGLK2, GLK2, GOLDEN2-like 2, GPRI2, GBF'S PRO-RICH REGION-INTERACTING FACTOR 2 |
0.236 |
0.371 |
| 139 |
248330_at |
AT5G52810
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.236 |
-0.057 |
| 140 |
249932_at |
AT5G22390
|
unknown |
0.235 |
0.051 |
| 141 |
261712_at |
AT1G32780
|
[GroES-like zinc-binding dehydrogenase family protein] |
0.235 |
0.136 |
| 142 |
251621_at |
AT3G57700
|
[Protein kinase superfamily protein] |
0.235 |
0.199 |
| 143 |
252501_at |
AT3G46880
|
unknown |
0.235 |
-0.477 |
| 144 |
252911_at |
AT4G39510
|
CYP96A12, cytochrome P450, family 96, subfamily A, polypeptide 12 |
0.235 |
0.279 |
| 145 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
0.234 |
0.201 |
| 146 |
259671_at |
AT1G52290
|
AtPERK15, proline-rich extensin-like receptor kinase 15, PERK15, proline-rich extensin-like receptor kinase 15 |
0.234 |
0.549 |
| 147 |
267380_at |
AT2G26170
|
CYP711A1, cytochrome P450, family 711, subfamily A, polypeptide 1, MAX1, MORE AXILLARY BRANCHES 1 |
0.234 |
0.286 |
| 148 |
257715_at |
AT3G12750
|
AtZIP1, ZIP1, zinc transporter 1 precursor |
0.232 |
0.384 |
| 149 |
253247_at |
AT4G34610
|
BLH6, BEL1-like homeodomain 6 |
0.231 |
0.194 |
| 150 |
252972_at |
AT4G38840
|
SAUR14, SMALL AUXIN UPREGULATED RNA 14 |
0.231 |
0.153 |
| 151 |
261807_at |
AT1G30515
|
unknown |
0.231 |
-0.021 |
| 152 |
263433_at |
AT2G22240
|
ATIPS2, INOSITOL 3-PHOSPHATE SYNTHASE 2, ATMIPS2, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 2, MIPS2, myo-inositol-1-phosphate synthase 2 |
0.231 |
0.427 |
| 153 |
246494_at |
AT5G16190
|
ATCSLA11, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE A11, CSLA11, cellulose synthase like A11 |
0.230 |
0.097 |
| 154 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
0.229 |
0.169 |
| 155 |
256073_at |
AT1G18100
|
E12A11, MFT, MOTHER OF FT AND TFL1 |
0.227 |
-0.146 |
| 156 |
261907_at |
AT1G65060
|
4CL3, 4-coumarate:CoA ligase 3 |
0.226 |
-0.098 |
| 157 |
247222_at |
AT5G64840
|
ABCF5, ATP-binding cassette F5, ATGCN5, general control non-repressible 5, GCN5, general control non-repressible 5 |
0.226 |
0.189 |
| 158 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.226 |
-0.051 |
| 159 |
251503_at |
AT3G59140
|
ABCC10, ATP-binding cassette C10, ATMRP14, multidrug resistance-associated protein 14, MRP14, multidrug resistance-associated protein 14 |
0.224 |
0.018 |
| 160 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
0.224 |
0.584 |
| 161 |
266391_at |
AT2G41290
|
SSL2, strictosidine synthase-like 2 |
0.224 |
0.247 |
| 162 |
253101_at |
AT4G37430
|
CYP81F1, CYTOCHROME P450 MONOOXYGENASE 81F1, CYP91A2, cytochrome P450, family 91, subfamily A, polypeptide 2 |
0.223 |
0.239 |
| 163 |
250926_at |
AT5G03555
|
AtNCS1, NCS1, nucleobase cation symporter 1 |
0.223 |
0.193 |
| 164 |
260451_at |
AT1G72360
|
AtERF73, ERF73, ethylene response factor 73, HRE1, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 1 |
0.222 |
0.390 |
| 165 |
266993_at |
AT2G39210
|
[Major facilitator superfamily protein] |
0.222 |
0.113 |
| 166 |
255627_at |
AT4G00955
|
unknown |
0.221 |
0.191 |
| 167 |
260466_at |
AT1G10900
|
[Phosphatidylinositol-4-phosphate 5-kinase family protein] |
0.221 |
-0.042 |
| 168 |
262882_at |
AT1G64900
|
CYP89, CYP89A2, cytochrome P450, family 89, subfamily A, polypeptide 2 |
0.221 |
-0.097 |
| 169 |
253394_at |
AT4G32770
|
ATSDX1, SUCROSE EXPORT DEFECTIVE 1, VTE1, VITAMIN E DEFICIENT 1 |
0.219 |
0.023 |
| 170 |
247406_at |
AT5G62920
|
ARR6, response regulator 6 |
0.219 |
-0.379 |
| 171 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
0.217 |
0.057 |
| 172 |
255591_at |
AT4G01630
|
ATEXP17, EXPANSIN 17, ATEXPA17, expansin A17, ATHEXP ALPHA 1.13, EXPA17, expansin A17 |
0.217 |
0.086 |
| 173 |
266765_at |
AT2G46860
|
AtPPa3, pyrophosphorylase 3, PPa3, pyrophosphorylase 3 |
0.217 |
-0.225 |
| 174 |
249727_at |
AT5G35490
|
ATMRU1, ARABIDOPSIS MTO 1 RESPONDING UP 1, MRU1, MTO 1 RESPONDING UP 1 |
0.216 |
0.291 |
| 175 |
253613_at |
AT4G30320
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.216 |
-0.055 |
| 176 |
267423_at |
AT2G35060
|
KUP11, K+ uptake permease 11 |
0.214 |
0.120 |
| 177 |
247467_at |
AT5G62130
|
[Per1-like family protein] |
0.214 |
0.320 |
| 178 |
251154_at |
AT3G63110
|
ATIPT3, isopentenyltransferase 3, IPT3, isopentenyltransferase 3 |
0.213 |
0.196 |
| 179 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.212 |
0.202 |
| 180 |
258890_at |
AT3G05690
|
ATHAP2B, HEME ACTIVATOR PROTEIN (YEAST) HOMOLOG 2B, HAP2B, HEME ACTIVATOR PROTEIN (YEAST) HOMOLOG 2B, NF-YA2, nuclear factor Y, subunit A2, UNE8, UNFERTILIZED EMBRYO SAC 8 |
0.212 |
0.236 |
| 181 |
264252_at |
AT1G09180
|
ATSAR1, SECRETION-ASSOCIATED RAS 1, ATSARA1A, secretion-associated RAS super family 1, SARA1A, secretion-associated RAS super family 1 |
0.212 |
0.051 |
| 182 |
245528_at |
AT4G15530
|
PPDK, pyruvate orthophosphate dikinase |
0.211 |
0.266 |
| 183 |
260601_at |
AT1G55910
|
ZIP11, zinc transporter 11 precursor |
0.211 |
0.158 |
| 184 |
247899_at |
AT5G57345
|
unknown |
0.211 |
0.139 |
| 185 |
260955_at |
AT1G06000
|
[UDP-Glycosyltransferase superfamily protein] |
0.210 |
-0.104 |
| 186 |
257185_at |
AT3G13100
|
ABCC7, ATP-binding cassette C7, ATMRP7, multidrug resistance-associated protein 7, MRP7, MRP7, multidrug resistance-associated protein 7 |
0.209 |
-0.045 |
| 187 |
256386_at |
AT1G66540
|
[Cytochrome P450 superfamily protein] |
0.209 |
-0.220 |
| 188 |
261016_at |
AT1G26560
|
BGLU40, beta glucosidase 40 |
0.209 |
0.090 |
| 189 |
249622_at |
AT5G37550
|
unknown |
0.209 |
0.027 |
| 190 |
257635_at |
AT3G26280
|
CYP71B4, cytochrome P450, family 71, subfamily B, polypeptide 4 |
0.209 |
0.127 |
| 191 |
249059_at |
AT5G44530
|
[Subtilase family protein] |
0.208 |
0.245 |
| 192 |
245479_at |
AT4G16140
|
[proline-rich family protein] |
0.207 |
0.348 |
| 193 |
247038_at |
AT5G67160
|
EPS1, ENHANCED PSEUDOMONAS SUSCEPTIBILTY 1 |
0.207 |
0.018 |
| 194 |
267535_at |
AT2G41940
|
ZFP8, zinc finger protein 8 |
0.206 |
0.239 |
| 195 |
246310_at |
AT3G51895
|
AST12, SULTR3;1, sulfate transporter 3;1 |
0.206 |
0.266 |
| 196 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
0.206 |
-0.331 |
| 197 |
264901_at |
AT1G23090
|
AST91, sulfate transporter 91, SULTR3;3 |
0.204 |
0.027 |
| 198 |
264696_at |
AT1G70230
|
AXY4, ALTERED XYLOGLUCAN 4, TBL27, TRICHOME BIREFRINGENCE-LIKE 27 |
0.204 |
0.352 |
| 199 |
258167_at |
AT3G21560
|
BRT1, Bright Trichomes 1, UGT84A2, UDP-glucosyl transferase 84A2 |
0.204 |
0.107 |
| 200 |
248686_at |
AT5G48540
|
[receptor-like protein kinase-related family protein] |
0.203 |
0.214 |
| 201 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
0.203 |
-0.157 |
| 202 |
267076_at |
AT2G41090
|
[Calcium-binding EF-hand family protein] |
0.202 |
0.016 |
| 203 |
246075_at |
AT5G20410
|
ATMGD2, ARABIDOPSIS THALIANA MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE 2, MGD2, monogalactosyldiacylglycerol synthase 2 |
0.201 |
0.393 |
| 204 |
260676_at |
AT1G19450
|
[Major facilitator superfamily protein] |
0.201 |
0.028 |
| 205 |
246888_at |
AT5G26270
|
unknown |
0.201 |
-0.063 |
| 206 |
256366_at |
AT1G66880
|
[Protein kinase superfamily protein] |
0.201 |
-0.002 |
| 207 |
254575_at |
AT4G19460
|
[UDP-Glycosyltransferase superfamily protein] |
0.200 |
0.117 |
| 208 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
0.200 |
0.114 |
| 209 |
250257_at |
AT5G13770
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.199 |
0.193 |
| 210 |
265722_at |
AT2G40100
|
LHCB4.3, light harvesting complex photosystem II |
0.197 |
-0.172 |
| 211 |
262577_at |
AT1G15290
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.197 |
0.174 |
| 212 |
248028_at |
AT5G55620
|
unknown |
0.196 |
0.037 |
| 213 |
254564_at |
AT4G19170
|
CCD4, carotenoid cleavage dioxygenase 4, NCED4, nine-cis-epoxycarotenoid dioxygenase 4 |
0.196 |
0.307 |
| 214 |
250646_at |
AT5G06720
|
ATPA2, peroxidase 2, AtPRX53, PA2, peroxidase 2, PRX53, peroxidase 53 |
0.196 |
0.089 |
| 215 |
264583_at |
AT1G05170
|
[Galactosyltransferase family protein] |
0.196 |
-0.058 |
| 216 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
0.195 |
-0.335 |
| 217 |
251454_at |
AT3G60080
|
[RING/U-box superfamily protein] |
0.195 |
0.222 |
| 218 |
246997_at |
AT5G67390
|
unknown |
0.194 |
0.118 |
| 219 |
266996_at |
AT2G34490
|
CYP710A2, cytochrome P450, family 710, subfamily A, polypeptide 2 |
0.194 |
0.160 |
| 220 |
249542_at |
AT5G38140
|
NF-YC12, nuclear factor Y, subunit C12 |
0.194 |
0.142 |
| 221 |
257297_at |
AT3G28040
|
[Leucine-rich receptor-like protein kinase family protein] |
0.194 |
0.141 |
| 222 |
256874_at |
AT3G26320
|
CYP71B36, cytochrome P450, family 71, subfamily B, polypeptide 36 |
0.193 |
-0.071 |
| 223 |
254693_at |
AT4G17880
|
MYC4 |
0.193 |
0.035 |
| 224 |
258315_at |
AT3G16175
|
[Thioesterase superfamily protein] |
0.193 |
0.314 |
| 225 |
245460_at |
AT4G16990
|
RLM3, RESISTANCE TO LEPTOSPHAERIA MACULANS 3 |
0.193 |
0.255 |
| 226 |
256446_at |
AT3G11110
|
[RING/U-box superfamily protein] |
0.192 |
0.260 |
| 227 |
266717_at |
AT2G46735
|
unknown |
0.192 |
0.204 |
| 228 |
251024_at |
AT5G02180
|
[Transmembrane amino acid transporter family protein] |
0.192 |
0.180 |
| 229 |
249214_at |
AT5G42720
|
[Glycosyl hydrolase family 17 protein] |
0.192 |
0.072 |
| 230 |
252917_at |
AT4G38960
|
BBX19, B-box domain protein 19 |
0.191 |
0.263 |
| 231 |
257916_at |
AT3G23210
|
bHLH34, basic Helix-Loop-Helix 34 |
0.190 |
0.084 |
| 232 |
264609_at |
AT1G04530
|
TPR4, tetratricopeptide repeat 4 |
0.190 |
0.066 |
| 233 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
-1.115 |
-0.714 |
| 234 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
-0.925 |
-0.306 |
| 235 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
-0.859 |
-0.324 |
| 236 |
263182_at |
AT1G05575
|
unknown |
-0.833 |
-0.024 |
| 237 |
253322_at |
AT4G33980
|
unknown |
-0.824 |
-0.303 |
| 238 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
-0.815 |
-0.208 |
| 239 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
-0.812 |
-0.528 |
| 240 |
256526_at |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
-0.780 |
-0.109 |
| 241 |
256442_at |
AT3G10930
|
unknown |
-0.776 |
-0.176 |
| 242 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.774 |
0.117 |
| 243 |
254074_at |
AT4G25490
|
ATCBF1, CBF1, C-repeat/DRE binding factor 1, DREB1B, DRE BINDING PROTEIN 1B |
-0.756 |
0.063 |
| 244 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
-0.731 |
-0.331 |
| 245 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
-0.722 |
-0.166 |
| 246 |
253643_at |
AT4G29780
|
unknown |
-0.702 |
0.113 |
| 247 |
266658_at |
AT2G25735
|
unknown |
-0.696 |
0.053 |
| 248 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
-0.683 |
-0.253 |
| 249 |
251977_at |
AT3G53250
|
SAUR57, SMALL AUXIN UPREGULATED RNA 57 |
-0.682 |
-0.359 |
| 250 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
-0.682 |
0.082 |
| 251 |
245749_at |
AT1G51090
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.672 |
0.267 |
| 252 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
-0.635 |
-0.009 |
| 253 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
-0.621 |
0.008 |
| 254 |
267036_at |
AT2G38465
|
unknown |
-0.613 |
-0.156 |
| 255 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
-0.608 |
-0.145 |
| 256 |
262452_at |
AT1G11210
|
unknown |
-0.607 |
0.278 |
| 257 |
260804_at |
AT1G78410
|
[VQ motif-containing protein] |
-0.598 |
-0.330 |
| 258 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
-0.595 |
0.064 |
| 259 |
248870_at |
AT5G46710
|
[PLATZ transcription factor family protein] |
-0.591 |
-0.299 |
| 260 |
247524_at |
AT5G61440
|
ACHT5, atypical CYS HIS rich thioredoxin 5 |
-0.585 |
-0.292 |
| 261 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
-0.580 |
-0.021 |
| 262 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
-0.577 |
0.060 |
| 263 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
-0.577 |
0.133 |
| 264 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
-0.558 |
0.049 |
| 265 |
258792_at |
AT3G04640
|
[glycine-rich protein] |
-0.555 |
-0.023 |
| 266 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
-0.554 |
-0.016 |
| 267 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
-0.550 |
-0.295 |
| 268 |
262307_at |
AT1G71000
|
[Chaperone DnaJ-domain superfamily protein] |
-0.544 |
-0.251 |
| 269 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
-0.542 |
-0.380 |
| 270 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
-0.540 |
0.059 |
| 271 |
253044_at |
AT4G37290
|
unknown |
-0.540 |
-0.276 |
| 272 |
246929_at |
AT5G25210
|
unknown |
-0.539 |
-0.114 |
| 273 |
260399_at |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
-0.537 |
-0.051 |
| 274 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
-0.535 |
-0.073 |
| 275 |
266841_at |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
-0.535 |
-0.673 |
| 276 |
260744_at |
AT1G15010
|
unknown |
-0.534 |
0.125 |
| 277 |
259992_at |
AT1G67970
|
AT-HSFA8, HSFA8, heat shock transcription factor A8 |
-0.528 |
-0.207 |
| 278 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
-0.523 |
-0.011 |
| 279 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
-0.522 |
0.277 |
| 280 |
248432_at |
AT5G51390
|
unknown |
-0.522 |
-0.280 |
| 281 |
246018_at |
AT5G10695
|
unknown |
-0.512 |
-0.022 |
| 282 |
260411_at |
AT1G69890
|
unknown |
-0.512 |
-0.002 |
| 283 |
259244_at |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
-0.511 |
-0.284 |
| 284 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
-0.510 |
-0.058 |
| 285 |
254954_at |
AT4G10910
|
unknown |
-0.508 |
-0.023 |
| 286 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.508 |
-0.108 |
| 287 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
-0.504 |
0.065 |
| 288 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
-0.501 |
0.060 |
| 289 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
-0.498 |
-0.150 |
| 290 |
264866_at |
AT1G24140
|
[Matrixin family protein] |
-0.497 |
-0.251 |
| 291 |
260688_at |
AT1G17665
|
unknown |
-0.493 |
-0.234 |
| 292 |
258225_at |
AT3G15630
|
unknown |
-0.487 |
-0.140 |
| 293 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
-0.487 |
0.093 |
| 294 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
-0.484 |
-0.271 |
| 295 |
248502_at |
AT5G50450
|
[HCP-like superfamily protein with MYND-type zinc finger] |
-0.484 |
-0.260 |
| 296 |
257670_at |
AT3G20340
|
unknown |
-0.480 |
-0.336 |
| 297 |
260748_at |
AT1G49210
|
[RING/U-box superfamily protein] |
-0.477 |
-0.086 |
| 298 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
-0.477 |
0.112 |
| 299 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
-0.474 |
-0.084 |
| 300 |
251494_at |
AT3G59350
|
[Protein kinase superfamily protein] |
-0.470 |
0.077 |
| 301 |
258742_at |
AT3G05800
|
AIF1, AtBS1(activation-tagged BRI1 suppressor 1)-interacting factor 1 |
-0.465 |
0.030 |
| 302 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
-0.460 |
-0.081 |
| 303 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
-0.459 |
-0.123 |
| 304 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
-0.458 |
-0.079 |
| 305 |
265511_at |
AT2G05540
|
[Glycine-rich protein family] |
-0.455 |
-0.014 |
| 306 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
-0.454 |
-0.982 |
| 307 |
265184_at |
AT1G23710
|
unknown |
-0.452 |
-0.062 |
| 308 |
261431_at |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
-0.448 |
-0.225 |
| 309 |
255733_at |
AT1G25400
|
unknown |
-0.446 |
-0.101 |
| 310 |
255381_at |
AT4G03510
|
ATRMA1, RMA1, RING membrane-anchor 1 |
-0.444 |
-0.135 |
| 311 |
263495_at |
AT2G42530
|
COR15B, cold regulated 15b |
-0.440 |
-0.034 |
| 312 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
-0.431 |
-0.231 |
| 313 |
267028_at |
AT2G38470
|
ATWRKY33, WRKY DNA-BINDING PROTEIN 33, WRKY33, WRKY DNA-binding protein 33 |
-0.428 |
-0.042 |
| 314 |
248744_at |
AT5G48250
|
BBX8, B-box domain protein 8 |
-0.426 |
-0.123 |
| 315 |
264774_at |
AT1G22890
|
unknown |
-0.425 |
0.020 |
| 316 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
-0.419 |
0.055 |
| 317 |
255753_at |
AT1G18570
|
AtMYB51, myb domain protein 51, BW51A, BW51B, HIG1, HIGH INDOLIC GLUCOSINOLATE 1, MYB51, myb domain protein 51 |
-0.416 |
0.250 |
| 318 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
-0.415 |
-0.174 |
| 319 |
265539_at |
AT2G15830
|
unknown |
-0.415 |
0.051 |
| 320 |
254193_at |
AT4G23870
|
unknown |
-0.411 |
0.108 |
| 321 |
265918_at |
AT2G15090
|
KCS8, 3-ketoacyl-CoA synthase 8 |
-0.411 |
0.060 |
| 322 |
256743_at |
AT3G29370
|
P1R3, P1R3 |
-0.407 |
-0.015 |
| 323 |
250649_at |
AT5G06690
|
WCRKC1, WCRKC thioredoxin 1 |
-0.407 |
-0.088 |
| 324 |
258436_at |
AT3G16720
|
ATL2, TOXICOS EN LEVADURA 2, TL2, TOXICOS EN LEVADURA 2 |
-0.402 |
0.006 |
| 325 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
-0.400 |
-0.034 |
| 326 |
260656_at |
AT1G19380
|
unknown |
-0.397 |
0.227 |
| 327 |
253292_at |
AT4G33985
|
unknown |
-0.392 |
-0.001 |
| 328 |
265725_at |
AT2G32030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.391 |
0.176 |
| 329 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
-0.391 |
-0.126 |
| 330 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
-0.388 |
-0.518 |
| 331 |
266259_at |
AT2G27830
|
unknown |
-0.384 |
-0.053 |
| 332 |
260527_at |
AT2G47270
|
UPB1, UPBEAT1 |
-0.381 |
-0.504 |
| 333 |
255607_at |
AT4G01130
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.381 |
0.041 |
| 334 |
265987_at |
AT2G24240
|
[BTB/POZ domain with WD40/YVTN repeat-like protein] |
-0.380 |
-0.050 |
| 335 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
-0.380 |
-0.144 |
| 336 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
-0.377 |
-0.200 |
| 337 |
251507_at |
AT3G59080
|
[Eukaryotic aspartyl protease family protein] |
-0.375 |
0.148 |
| 338 |
252483_at |
AT3G46600
|
[GRAS family transcription factor] |
-0.371 |
-0.108 |
| 339 |
255742_at |
AT1G25560
|
AtTEM1, EDF1, ETHYLENE RESPONSE DNA BINDING FACTOR 1, TEM1, TEMPRANILLO 1 |
-0.370 |
0.051 |
| 340 |
246200_at |
AT4G37240
|
unknown |
-0.367 |
-0.018 |
| 341 |
247047_at |
AT5G66650
|
unknown |
-0.365 |
0.175 |
| 342 |
266590_at |
AT2G46240
|
ATBAG6, ARABIDOPSIS THALIANA BCL-2-ASSOCIATED ATHANOGENE 6, BAG6, BCL-2-associated athanogene 6 |
-0.365 |
-0.344 |
| 343 |
259979_at |
AT1G76600
|
unknown |
-0.364 |
-0.259 |
| 344 |
261453_at |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
-0.358 |
0.153 |
| 345 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
-0.358 |
-0.054 |
| 346 |
267083_at |
AT2G41100
|
ATCAL4, ARABIDOPSIS THALIANA CALMODULIN LIKE 4, TCH3, TOUCH 3 |
-0.356 |
0.025 |
| 347 |
252539_at |
AT3G45730
|
unknown |
-0.354 |
-0.006 |
| 348 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
-0.353 |
0.250 |
| 349 |
267364_at |
AT2G40080
|
ELF4, EARLY FLOWERING 4 |
-0.353 |
-0.397 |
| 350 |
265387_at |
AT2G20670
|
unknown |
-0.352 |
-0.174 |
| 351 |
247708_at |
AT5G59550
|
unknown |
-0.351 |
0.007 |
| 352 |
266396_at |
AT2G38790
|
unknown |
-0.351 |
-0.000 |
| 353 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
-0.351 |
-0.042 |
| 354 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
-0.350 |
0.155 |
| 355 |
267254_at |
AT2G23030
|
SNRK2-9, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-9, SNRK2.9, SNF1-related protein kinase 2.9 |
-0.350 |
-0.175 |
| 356 |
261470_at |
AT1G28370
|
ATERF11, ERF DOMAIN PROTEIN 11, ERF11, ERF domain protein 11 |
-0.349 |
0.142 |
| 357 |
254300_at |
AT4G22780
|
ACR7, ACT domain repeat 7 |
-0.348 |
0.026 |
| 358 |
264045_at |
AT2G22450
|
AtRIBA2, RIBA2, homolog of ribA 2 |
-0.346 |
-0.108 |
| 359 |
267381_at |
AT2G26190
|
[calmodulin-binding family protein] |
-0.341 |
0.015 |
| 360 |
256755_at |
AT3G25600
|
[Calcium-binding EF-hand family protein] |
-0.340 |
0.117 |
| 361 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
-0.339 |
0.089 |
| 362 |
252053_at |
AT3G52400
|
ATSYP122, SYP122, syntaxin of plants 122 |
-0.339 |
0.127 |
| 363 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
-0.339 |
-0.096 |
| 364 |
255931_at |
AT1G12710
|
AtPP2-A12, phloem protein 2-A12, PP2-A12, phloem protein 2-A12 |
-0.338 |
-0.075 |
| 365 |
247525_at |
AT5G61380
|
APRR1, AtTOC1, TIMING OF CAB EXPRESSION 1, PRR1, PSEUDO-RESPONSE REGULATOR 1, TOC1, TIMING OF CAB EXPRESSION 1 |
-0.338 |
-0.084 |
| 366 |
254922_at |
AT4G11370
|
RHA1A, RING-H2 finger A1A |
-0.337 |
-0.409 |
| 367 |
257022_at |
AT3G19580
|
AZF2, zinc-finger protein 2, ZF2, zinc-finger protein 2 |
-0.337 |
-0.140 |
| 368 |
263379_at |
AT2G40140
|
ATSZF2, CZF1, SZF2, (SALT-INDUCIBLE ZINC FINGER 2, ZFAR1 |
-0.336 |
0.001 |
| 369 |
255511_at |
AT4G02075
|
PIT1, pitchoun 1 |
-0.335 |
-0.353 |
| 370 |
247137_at |
AT5G66210
|
CPK28, calcium-dependent protein kinase 28 |
-0.334 |
0.190 |
| 371 |
259694_at |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
-0.333 |
-0.085 |
| 372 |
260227_at |
AT1G74450
|
unknown |
-0.331 |
0.117 |
| 373 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
-0.329 |
0.118 |
| 374 |
256418_at |
AT3G06160
|
[AP2/B3-like transcriptional factor family protein] |
-0.327 |
-0.106 |
| 375 |
266278_at |
AT2G29300
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.325 |
-0.263 |
| 376 |
253737_at |
AT4G28703
|
[RmlC-like cupins superfamily protein] |
-0.325 |
-0.038 |
| 377 |
264482_at |
AT1G77210
|
AtSTP14, sugar transport protein 14, STP14, sugar transport protein 14 |
-0.323 |
-0.011 |
| 378 |
248392_at |
AT5G52050
|
[MATE efflux family protein] |
-0.321 |
0.077 |
| 379 |
250279_at |
AT5G13200
|
[GRAM domain family protein] |
-0.320 |
0.109 |
| 380 |
262092_at |
AT1G56150
|
SAUR71, SMALL AUXIN UPREGULATED 71 |
-0.319 |
-0.101 |
| 381 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.318 |
-0.036 |
| 382 |
258939_at |
AT3G10020
|
unknown |
-0.317 |
-0.215 |
| 383 |
251631_at |
AT3G57430
|
OTP84, ORGANELLE TRANSCRIPT PROCESSING 84 |
-0.317 |
-0.082 |
| 384 |
246253_at |
AT4G37260
|
ATMYB73, MYB73, myb domain protein 73 |
-0.317 |
-0.102 |
| 385 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
-0.316 |
-0.048 |
| 386 |
247867_at |
AT5G57630
|
CIPK21, CBL-interacting protein kinase 21, SnRK3.4, SNF1-RELATED PROTEIN KINASE 3.4 |
-0.316 |
-0.427 |
| 387 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
-0.314 |
0.173 |
| 388 |
248793_at |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
-0.314 |
-0.440 |
| 389 |
253421_at |
AT4G32340
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.313 |
-0.079 |
| 390 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
-0.313 |
0.016 |
| 391 |
266983_at |
AT2G39400
|
[alpha/beta-Hydrolases superfamily protein] |
-0.312 |
0.008 |
| 392 |
256503_at |
AT1G75250
|
ATRL6, RAD-like 6, RL6, RAD-like 6, RSM3, RADIALIS-LIKE SANT/MYB 3 |
-0.311 |
-0.072 |
| 393 |
254178_at |
AT4G23880
|
unknown |
-0.310 |
0.068 |
| 394 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
-0.310 |
-0.185 |
| 395 |
259211_at |
AT3G09020
|
[alpha 1,4-glycosyltransferase family protein] |
-0.309 |
-0.137 |
| 396 |
264787_at |
AT2G17840
|
ERD7, EARLY-RESPONSIVE TO DEHYDRATION 7 |
-0.309 |
0.041 |
| 397 |
264389_at |
AT1G11960
|
[ERD (early-responsive to dehydration stress) family protein] |
-0.307 |
0.052 |
| 398 |
249872_at |
AT5G23130
|
[Peptidoglycan-binding LysM domain-containing protein] |
-0.306 |
-0.032 |
| 399 |
252278_at |
AT3G49530
|
ANAC062, NAC domain containing protein 62, NAC062, NAC domain containing protein 62, NTL6, NTM1 (NAC WITH TRANSMEMBRANE MOTIF 1)-LIKE 6 |
-0.306 |
-0.033 |
| 400 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
-0.305 |
-0.371 |
| 401 |
259595_at |
AT1G28050
|
BBX13, B-box domain protein 13 |
-0.305 |
-0.111 |
| 402 |
249234_at |
AT5G42200
|
[RING/U-box superfamily protein] |
-0.304 |
-0.045 |
| 403 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
-0.304 |
-0.038 |
| 404 |
247786_at |
AT5G58600
|
PMR5, POWDERY MILDEW RESISTANT 5, TBL44, TRICHOME BIREFRINGENCE-LIKE 44 |
-0.303 |
0.096 |
| 405 |
247348_at |
AT5G63810
|
AtBGAL10, Arabidopsis thaliana beta-galactosidase 10, BGAL10, beta-galactosidase 10 |
-0.303 |
-0.157 |
| 406 |
263800_at |
AT2G24600
|
[Ankyrin repeat family protein] |
-0.302 |
0.105 |
| 407 |
262164_at |
AT1G78070
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.302 |
0.243 |
| 408 |
267460_at |
AT2G33810
|
SPL3, squamosa promoter binding protein-like 3 |
-0.298 |
0.021 |
| 409 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.297 |
-0.326 |
| 410 |
247013_at |
AT5G67480
|
ATBT4, BT4, BTB and TAZ domain protein 4 |
-0.297 |
-0.083 |
| 411 |
254592_at |
AT4G18880
|
AT-HSFA4A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A4A, HSF A4A, heat shock transcription factor A4A |
-0.295 |
-0.128 |
| 412 |
267048_at |
AT2G34200
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-0.295 |
-0.126 |
| 413 |
245906_at |
AT5G11070
|
unknown |
-0.293 |
-0.133 |
| 414 |
250550_at |
AT5G07870
|
[HXXXD-type acyl-transferase family protein] |
-0.291 |
-0.238 |
| 415 |
246270_at |
AT4G36500
|
unknown |
-0.290 |
0.009 |
| 416 |
256455_at |
AT1G75190
|
unknown |
-0.290 |
0.001 |
| 417 |
267293_at |
AT2G23810
|
TET8, tetraspanin8 |
-0.290 |
-0.001 |
| 418 |
260146_at |
AT1G52770
|
[Phototropic-responsive NPH3 family protein] |
-0.290 |
-0.470 |
| 419 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
-0.288 |
-0.056 |
| 420 |
261265_at |
AT1G26800
|
[RING/U-box superfamily protein] |
-0.288 |
-0.342 |
| 421 |
253872_at |
AT4G27410
|
ANAC072, Arabidopsis NAC domain containing protein 72, RD26, RESPONSIVE TO DESICCATION 26 |
-0.287 |
-0.437 |
| 422 |
262236_at |
AT1G48330
|
unknown |
-0.287 |
-0.232 |
| 423 |
247848_at |
AT5G58120
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.286 |
0.196 |
| 424 |
256420_at |
AT1G33475
|
[SNARE-like superfamily protein] |
-0.286 |
-0.005 |
| 425 |
253323_at |
AT4G33920
|
APD5, Arabidopsis Pp2c clade D 5 |
-0.285 |
0.116 |
| 426 |
255844_at |
AT2G33580
|
LYK5, LysM-containing receptor-like kinase 5 |
-0.285 |
0.202 |
| 427 |
248821_at |
AT5G47070
|
[Protein kinase superfamily protein] |
-0.285 |
-0.063 |
| 428 |
256114_at |
AT1G16850
|
unknown |
-0.283 |
0.392 |
| 429 |
250676_at |
AT5G06320
|
NHL3, NDR1/HIN1-like 3 |
-0.283 |
0.054 |
| 430 |
266825_at |
AT2G22890
|
[Kua-ubiquitin conjugating enzyme hybrid localisation domain] |
-0.283 |
0.009 |
| 431 |
262892_at |
AT1G79440
|
ALDH5F1, aldehyde dehydrogenase 5F1, ENF1, ENLARGED FIL EXPRESSION DOMAIN 1, SSADH, SUCCINIC SEMIALDEHYDE DEHYDROGENASE, SSADH1, SUCCINIC SEMIALDEHYDE DEHYDROGENASE 1 |
-0.283 |
-0.092 |
| 432 |
253331_at |
AT4G33490
|
[Eukaryotic aspartyl protease family protein] |
-0.282 |
-0.099 |
| 433 |
255939_at |
AT1G12730
|
[GPI transamidase subunit PIG-U] |
-0.282 |
-0.296 |
| 434 |
250194_at |
AT5G14550
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
-0.281 |
-0.105 |
| 435 |
264211_at |
AT1G22770
|
FB, GI, GIGANTEA |
-0.281 |
-0.019 |
| 436 |
253813_at |
AT4G28150
|
unknown |
-0.281 |
-0.196 |
| 437 |
247177_at |
AT5G65300
|
unknown |
-0.280 |
0.056 |
| 438 |
263919_at |
AT2G36470
|
unknown |
-0.280 |
-0.116 |
| 439 |
247240_at |
AT5G64660
|
ATCMPG2, CMPG2, CYS, MET, PRO, and GLY protein 2 |
-0.275 |
0.142 |
| 440 |
251636_at |
AT3G57530
|
ATCPK32, CDPK32, CPK32, calcium-dependent protein kinase 32 |
-0.274 |
0.063 |
| 441 |
261177_at |
AT1G04770
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.274 |
-0.115 |
| 442 |
249719_at |
AT5G35735
|
[Auxin-responsive family protein] |
-0.274 |
-0.033 |
| 443 |
249746_at |
AT5G24590
|
ANAC091, Arabidopsis NAC domain containing protein 91, TIP, TCV-interacting protein |
-0.271 |
0.033 |
| 444 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
-0.270 |
0.261 |
| 445 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
-0.267 |
-0.095 |
| 446 |
254839_at |
AT4G12400
|
Hop3, Hop3 |
-0.267 |
-0.604 |
| 447 |
249472_at |
AT5G39210
|
CRR7, CHLORORESPIRATORY REDUCTION 7 |
-0.267 |
-0.114 |
| 448 |
259015_at |
AT3G07350
|
unknown |
-0.265 |
-0.204 |
| 449 |
251774_at |
AT3G55840
|
[Hs1pro-1 protein] |
-0.265 |
-0.031 |
| 450 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
-0.264 |
0.027 |
| 451 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
-0.264 |
0.203 |
| 452 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
-0.263 |
-0.099 |
| 453 |
251603_at |
AT3G57760
|
[Protein kinase superfamily protein] |
-0.263 |
-0.158 |
| 454 |
264511_at |
AT1G09350
|
AtGolS3, galactinol synthase 3, GolS3, galactinol synthase 3 |
-0.263 |
0.225 |
| 455 |
254158_at |
AT4G24380
|
[INVOLVED IN: 10-formyltetrahydrofolate biosynthetic process, folic acid and derivative biosynthetic process] |
-0.263 |
-0.101 |
| 456 |
256715_at |
AT2G34090
|
MEE18, maternal effect embryo arrest 18 |
-0.262 |
-0.005 |
| 457 |
263475_at |
AT2G31945
|
unknown |
-0.262 |
-0.191 |
| 458 |
254580_at |
AT4G19390
|
[Uncharacterised protein family (UPF0114)] |
-0.262 |
-0.021 |
| 459 |
259312_at |
AT3G05200
|
ATL6, Arabidopsis toxicos en levadura 6 |
-0.262 |
0.046 |
| 460 |
245247_at |
AT4G17230
|
SCL13, SCARECROW-like 13 |
-0.260 |
-0.119 |
| 461 |
255064_at |
AT4G08950
|
EXO, EXORDIUM |
-0.258 |
0.132 |
| 462 |
261613_at |
AT1G49720
|
ABF1, abscisic acid responsive element-binding factor 1, AtABF1 |
-0.257 |
-0.065 |
| 463 |
245699_at |
AT5G04250
|
[Cysteine proteinases superfamily protein] |
-0.257 |
-0.119 |