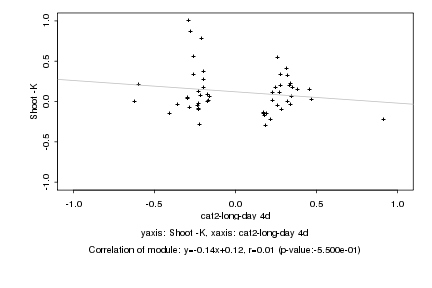
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
cat2-long-day 4d |
Log2 signal ratio
Shoot -K |
| 1 |
251772_at |
AT3G55920
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
0.911 |
-0.220 |
| 2 |
262719_at |
AT1G43590
|
unknown |
0.464 |
0.026 |
| 3 |
257670_at |
AT3G20340
|
unknown |
0.456 |
0.150 |
| 4 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
0.379 |
0.155 |
| 5 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.352 |
0.179 |
| 6 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.346 |
0.067 |
| 7 |
253281_at |
AT4G34138
|
UGT73B1, UDP-glucosyl transferase 73B1 |
0.335 |
-0.036 |
| 8 |
255543_at |
AT4G01870
|
[tolB protein-related] |
0.334 |
0.233 |
| 9 |
248037_at |
AT5G55930
|
ATOPT1, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 1, OPT1, oligopeptide transporter 1 |
0.332 |
0.203 |
| 10 |
254384_at |
AT4G21870
|
[HSP20-like chaperones superfamily protein] |
0.321 |
0.009 |
| 11 |
265170_at |
AT1G23730
|
ATBCA3, BETA CARBONIC ANHYDRASE 3, BCA3, beta carbonic anhydrase 3 |
0.320 |
0.328 |
| 12 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.310 |
0.419 |
| 13 |
262196_at |
AT1G77870
|
MUB5, membrane-anchored ubiquitin-fold protein 5 precursor |
0.282 |
-0.095 |
| 14 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.277 |
0.346 |
| 15 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
0.274 |
0.201 |
| 16 |
246195_at |
AT4G36410
|
UBC17, ubiquitin-conjugating enzyme 17 |
0.270 |
0.124 |
| 17 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.258 |
0.550 |
| 18 |
255732_at |
AT1G25450
|
CER60, ECERIFERUM 60, KCS5, 3-ketoacyl-CoA synthase 5 |
0.257 |
-0.049 |
| 19 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
0.245 |
0.185 |
| 20 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
0.226 |
0.020 |
| 21 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
0.226 |
0.114 |
| 22 |
251584_at |
AT3G58620
|
TTL4, tetratricopetide-repeat thioredoxin-like 4 |
0.214 |
-0.217 |
| 23 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
0.187 |
-0.142 |
| 24 |
254609_at |
AT4G18970
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.185 |
-0.286 |
| 25 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
0.174 |
-0.168 |
| 26 |
261548_at |
AT1G63480
|
AHL12, AT-hook motif nuclear localized protein 12 |
0.172 |
-0.135 |
| 27 |
266805_at |
AT2G30010
|
TBL45, TRICHOME BIREFRINGENCE-LIKE 45 |
0.168 |
-0.147 |
| 28 |
253174_at |
AT4G35090
|
CAT2, catalase 2 |
-0.627 |
0.008 |
| 29 |
251743_at |
AT3G55890
|
[Yippee family putative zinc-binding protein] |
-0.604 |
0.222 |
| 30 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
-0.411 |
-0.143 |
| 31 |
250485_at |
AT5G09990
|
PROPEP5, elicitor peptide 5 precursor |
-0.363 |
-0.025 |
| 32 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
-0.302 |
0.049 |
| 33 |
245384_at |
AT4G16790
|
[hydroxyproline-rich glycoprotein family protein] |
-0.301 |
0.053 |
| 34 |
248333_at |
AT5G52390
|
[PAR1 protein] |
-0.294 |
1.017 |
| 35 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.285 |
-0.069 |
| 36 |
249626_at |
AT5G37540
|
[Eukaryotic aspartyl protease family protein] |
-0.282 |
0.873 |
| 37 |
256114_at |
AT1G16850
|
unknown |
-0.263 |
0.560 |
| 38 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
-0.263 |
0.347 |
| 39 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
-0.239 |
-0.039 |
| 40 |
261881_at |
AT1G80760
|
NIP6, NIP6;1, NOD26-like intrinsic protein 6;1, NLM7 |
-0.231 |
-0.077 |
| 41 |
255575_at |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
-0.231 |
0.126 |
| 42 |
260805_at |
AT1G78320
|
ATGSTU23, glutathione S-transferase TAU 23, GSTU23, glutathione S-transferase TAU 23 |
-0.230 |
-0.093 |
| 43 |
257678_at |
AT3G20420
|
ATRTL2, RNASEIII-LIKE 2, RTL2, RNAse THREE-like protein 2 |
-0.230 |
-0.014 |
| 44 |
260380_at |
AT1G73870
|
BBX16, B-box domain protein 16, COL7, CONSTANS-LIKE 7 |
-0.228 |
-0.276 |
| 45 |
254735_at |
AT4G13810
|
AtRLP47, receptor like protein 47, RLP47, receptor like protein 47 |
-0.217 |
0.079 |
| 46 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
-0.211 |
0.784 |
| 47 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
-0.203 |
0.376 |
| 48 |
256529_at |
AT1G33260
|
[Protein kinase superfamily protein] |
-0.201 |
0.185 |
| 49 |
253113_at |
AT4G35985
|
[Senescence/dehydration-associated protein-related] |
-0.198 |
0.274 |
| 50 |
252083_at |
AT3G51960
|
ATBZIP24, basic leucine zipper 24, BZIP24, basic leucine zipper 24 |
-0.174 |
0.006 |
| 51 |
247358_at |
AT5G63580
|
ATFLS2, FLS2, flavonol synthase 2 |
-0.173 |
0.088 |
| 52 |
265978_at |
AT2G11140
|
[copia-like retrotransposon family, has a 1.0e-71 P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
-0.171 |
0.024 |
| 53 |
261212_at |
AT1G12880
|
atnudt12, nudix hydrolase homolog 12, NUDT12, nudix hydrolase homolog 12 |
-0.162 |
0.074 |