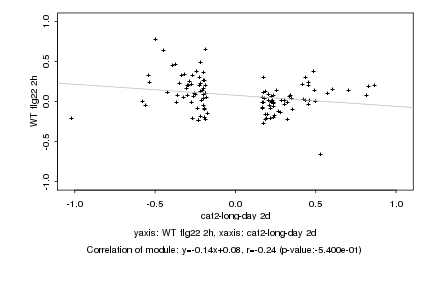
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
cat2-long-day 2d |
Log2 signal ratio
WT flg22 2h |
| 1 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.865 |
0.208 |
| 2 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.826 |
0.194 |
| 3 |
251772_at |
AT3G55920
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
0.813 |
0.087 |
| 4 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.702 |
0.150 |
| 5 |
246464_at |
AT5G16980
|
[Zinc-binding dehydrogenase family protein] |
0.601 |
0.161 |
| 6 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.571 |
0.108 |
| 7 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
0.529 |
-0.661 |
| 8 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
0.497 |
0.002 |
| 9 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.490 |
0.150 |
| 10 |
250152_at |
AT5G15120
|
unknown |
0.485 |
0.382 |
| 11 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
0.457 |
0.022 |
| 12 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.452 |
-0.034 |
| 13 |
260522_x_at |
AT2G41730
|
unknown |
0.450 |
0.206 |
| 14 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
0.450 |
0.248 |
| 15 |
253281_at |
AT4G34138
|
UGT73B1, UDP-glucosyl transferase 73B1 |
0.435 |
0.024 |
| 16 |
257670_at |
AT3G20340
|
unknown |
0.434 |
0.304 |
| 17 |
263073_at |
AT2G17500
|
[Auxin efflux carrier family protein] |
0.418 |
0.029 |
| 18 |
266752_at |
AT2G47000
|
ABCB4, ATP-binding cassette B4, AtABCB4, ATPGP4, ARABIDOPSIS P-GLYCOPROTEIN 4, MDR4, MULTIDRUG RESISTANCE 4, PGP4, P-GLYCOPROTEIN 4 |
0.414 |
0.221 |
| 19 |
262719_at |
AT1G43590
|
unknown |
0.350 |
-0.098 |
| 20 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
0.344 |
0.041 |
| 21 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.337 |
0.076 |
| 22 |
262935_at |
AT1G79410
|
AtOCT5, organic cation/carnitine transporter5, OCT5, organic cation/carnitine transporter5 |
0.334 |
0.064 |
| 23 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
0.321 |
-0.006 |
| 24 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
0.319 |
-0.215 |
| 25 |
263046_at |
AT2G05380
|
GRP3S, glycine-rich protein 3 short isoform |
0.305 |
0.017 |
| 26 |
246403_at |
AT1G57590
|
[Pectinacetylesterase family protein] |
0.302 |
-0.037 |
| 27 |
257547_at |
AT3G13000
|
unknown |
0.283 |
0.014 |
| 28 |
258209_at |
AT3G14060
|
unknown |
0.280 |
-0.125 |
| 29 |
250365_at |
AT5G11410
|
[Protein kinase superfamily protein] |
0.262 |
-0.114 |
| 30 |
248607_at |
AT5G49480
|
ATCP1, Ca2+-binding protein 1, CP1, Ca2+-binding protein 1 |
0.253 |
0.145 |
| 31 |
248801_at |
AT5G47370
|
HAT2 |
0.243 |
-0.163 |
| 32 |
267266_at |
AT2G23150
|
ATNRAMP3, NRAMP3, natural resistance-associated macrophage protein 3 |
0.236 |
-0.052 |
| 33 |
267349_at |
AT2G40010
|
[Ribosomal protein L10 family protein] |
0.234 |
-0.006 |
| 34 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.232 |
0.083 |
| 35 |
261636_at |
AT1G50110
|
[D-aminoacid aminotransferase-like PLP-dependent enzymes superfamily protein] |
0.232 |
-0.013 |
| 36 |
254384_at |
AT4G21870
|
[HSP20-like chaperones superfamily protein] |
0.231 |
-0.188 |
| 37 |
247109_at |
AT5G65870
|
ATPSK5, phytosulfokine 5 precursor, PSK5, PSK5, phytosulfokine 5 precursor |
0.229 |
0.022 |
| 38 |
264025_at |
AT2G21050
|
LAX2, like AUXIN RESISTANT 2 |
0.220 |
0.073 |
| 39 |
248037_at |
AT5G55930
|
ATOPT1, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 1, OPT1, oligopeptide transporter 1 |
0.220 |
0.011 |
| 40 |
257900_at |
AT3G28420
|
[Putative membrane lipoprotein] |
0.220 |
-0.006 |
| 41 |
262196_at |
AT1G77870
|
MUB5, membrane-anchored ubiquitin-fold protein 5 precursor |
0.218 |
-0.085 |
| 42 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
0.213 |
-0.210 |
| 43 |
247488_at |
AT5G61820
|
unknown |
0.208 |
-0.041 |
| 44 |
245277_at |
AT4G15550
|
IAGLU, indole-3-acetate beta-D-glucosyltransferase |
0.200 |
0.017 |
| 45 |
263515_at |
AT2G21640
|
unknown |
0.200 |
0.097 |
| 46 |
245362_at |
AT4G17460
|
HAT1, JAB, JAIBA |
0.199 |
-0.159 |
| 47 |
258033_at |
AT3G21250
|
ABCC8, ATP-binding cassette C8, ATMRP6, ARABIDOPSIS THALIANA MULTIDRUG RESISTANCE-ASSOCIATED PROTEIN 6, MRP6, multidrug resistance-associated protein 6 |
0.189 |
-0.206 |
| 48 |
266778_at |
AT2G29090
|
CYP707A2, cytochrome P450, family 707, subfamily A, polypeptide 2 |
0.187 |
-0.220 |
| 49 |
251584_at |
AT3G58620
|
TTL4, tetratricopetide-repeat thioredoxin-like 4 |
0.182 |
0.128 |
| 50 |
251235_at |
AT3G62860
|
[alpha/beta-Hydrolases superfamily protein] |
0.182 |
-0.150 |
| 51 |
246463_at |
AT5G16970
|
AER, alkenal reductase, AT-AER, alkenal reductase |
0.179 |
0.047 |
| 52 |
261564_at |
AT1G01720
|
ANAC002, Arabidopsis NAC domain containing protein 2, ATAF1 |
0.174 |
0.303 |
| 53 |
245692_at |
AT5G04150
|
BHLH101 |
0.171 |
-0.011 |
| 54 |
266296_at |
AT2G29420
|
ATGSTU7, glutathione S-transferase tau 7, GST25, GLUTATHIONE S-TRANSFERASE 25, GSTU7, glutathione S-transferase tau 7 |
0.169 |
0.114 |
| 55 |
263473_at |
AT2G31750
|
UGT74D1, UDP-glucosyl transferase 74D1 |
0.169 |
-0.265 |
| 56 |
267319_at |
AT2G34660
|
ABCC2, ATP-binding cassette C2, AtABCC2, Arabidopsis thaliana ATP-binding cassette C2, ATMRP2, multidrug resistance-associated protein 2, EST4, MRP2, multidrug resistance-associated protein 2 |
0.168 |
-0.006 |
| 57 |
251039_at |
AT5G02020
|
SIS, Salt Induced Serine rich |
0.167 |
0.056 |
| 58 |
263002_at |
AT1G54200
|
unknown |
0.165 |
-0.064 |
| 59 |
251317_at |
AT3G61490
|
[Pectin lyase-like superfamily protein] |
0.164 |
-0.081 |
| 60 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
-1.025 |
-0.205 |
| 61 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
-0.579 |
0.004 |
| 62 |
253174_at |
AT4G35090
|
CAT2, catalase 2 |
-0.560 |
-0.041 |
| 63 |
264648_at |
AT1G09080
|
BIP3, binding protein 3 |
-0.546 |
0.332 |
| 64 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
-0.539 |
0.248 |
| 65 |
249896_at |
AT5G22530
|
unknown |
-0.500 |
0.783 |
| 66 |
260904_at |
AT1G02450
|
NIMIN-1, NIMIN1, NIM1-interacting 1 |
-0.448 |
0.644 |
| 67 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
-0.428 |
0.121 |
| 68 |
252200_at |
AT3G50280
|
[HXXXD-type acyl-transferase family protein] |
-0.425 |
0.116 |
| 69 |
262671_at |
AT1G76040
|
CPK29, calcium-dependent protein kinase 29 |
-0.395 |
0.460 |
| 70 |
252060_at |
AT3G52430
|
ATPAD4, ARABIDOPSIS PHYTOALEXIN DEFICIENT 4, PAD4, PHYTOALEXIN DEFICIENT 4 |
-0.376 |
0.465 |
| 71 |
247224_at |
AT5G65080
|
AGL68, AGAMOUS-like 68, MAF5, MADS AFFECTING FLOWERING 5 |
-0.369 |
-0.002 |
| 72 |
249940_at |
AT5G22380
|
anac090, NAC domain containing protein 90, NAC090, NAC domain containing protein 90 |
-0.364 |
0.085 |
| 73 |
252403_at |
AT3G48080
|
[alpha/beta-Hydrolases superfamily protein] |
-0.349 |
0.232 |
| 74 |
252921_at |
AT4G39030
|
EDS5, ENHANCED DISEASE SUSCEPTIBILITY 5, SCORD3, susceptible to coronatine-deficient Pst DC3000 3, SID1, SALICYLIC ACID INDUCTION DEFICIENT 1 |
-0.336 |
0.332 |
| 75 |
263852_at |
AT2G04450
|
ATNUDT6, nudix hydrolase homolog 6, ATNUDX6, Arabidopsis thaliana nucleoside diphosphate linked to some moiety X 6, NUDT6, nudix hydrolase homolog 6, NUDX6, nucleoside diphosphates linked to some moiety X 6 |
-0.327 |
0.055 |
| 76 |
256526_at |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
-0.320 |
0.341 |
| 77 |
254243_at |
AT4G23210
|
CRK13, cysteine-rich RLK (RECEPTOR-like protein kinase) 13 |
-0.305 |
0.167 |
| 78 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
-0.300 |
0.207 |
| 79 |
247223_at |
AT5G65070
|
AGL69, AGAMOUS-like 69, FCL4, MAF4, MADS AFFECTING FLOWERING 4 |
-0.300 |
0.085 |
| 80 |
248916_at |
AT5G45840
|
[Leucine-rich repeat protein kinase family protein] |
-0.294 |
0.210 |
| 81 |
254252_at |
AT4G23310
|
CRK23, cysteine-rich RLK (RECEPTOR-like protein kinase) 23 |
-0.287 |
0.257 |
| 82 |
267380_at |
AT2G26170
|
CYP711A1, cytochrome P450, family 711, subfamily A, polypeptide 1, MAX1, MORE AXILLARY BRANCHES 1 |
-0.276 |
-0.012 |
| 83 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
-0.275 |
0.218 |
| 84 |
251633_at |
AT3G57460
|
[catalytics] |
-0.271 |
0.333 |
| 85 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
-0.270 |
-0.209 |
| 86 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
-0.263 |
0.064 |
| 87 |
252114_at |
AT3G51450
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.256 |
0.106 |
| 88 |
258445_at |
AT3G22400
|
ATLOX5, Arabidopsis thaliana lipoxygenase 5, LOX5 |
-0.250 |
0.100 |
| 89 |
262177_at |
AT1G74710
|
ATICS1, ARABIDOPSIS ISOCHORISMATE SYNTHASE 1, EDS16, ENHANCED DISEASE SUSCEPTIBILITY TO ERYSIPHE ORONTII 16, ICS1, ISOCHORISMATE SYNTHASE 1, SID2, SALICYLIC ACID INDUCTION DEFICIENT 2 |
-0.246 |
0.387 |
| 90 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
-0.237 |
-0.081 |
| 91 |
260774_at |
AT1G78290
|
SNRK2-8, SNF1-RELATED PROTEIN KINASE 2-8, SNRK2.8, SNF1-RELATED PROTEIN KINASE 2.8, SRK2C, SNF1-RELATED PROTEIN KINASE 2C |
-0.233 |
-0.237 |
| 92 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
-0.233 |
-0.228 |
| 93 |
252684_at |
AT3G44400
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.228 |
0.309 |
| 94 |
255319_at |
AT4G04220
|
AtRLP46, receptor like protein 46, RLP46, receptor like protein 46 |
-0.227 |
0.204 |
| 95 |
250994_at |
AT5G02490
|
AtHsp70-2, Hsp70-2 |
-0.221 |
0.233 |
| 96 |
259383_at |
AT3G16470
|
JAL35, jacalin-related lectin 35, JR1, JASMONATE RESPONSIVE 1 |
-0.221 |
0.130 |
| 97 |
259640_at |
AT1G52400
|
ATBG1, A. THALIANA BETA-GLUCOSIDASE 1, BGL1, BETA-GLUCOSIDASE HOMOLOG 1, BGLU18, beta glucosidase 18 |
-0.220 |
-0.187 |
| 98 |
264867_at |
AT1G24150
|
ATFH4, FORMIN HOMOLOGUE 4, FH4, formin homologue 4 |
-0.218 |
0.491 |
| 99 |
261826_at |
AT1G11580
|
ATPMEPCRA, PMEPCRA, methylesterase PCR A |
-0.217 |
0.019 |
| 100 |
261167_at |
AT1G04980
|
ATPDI10, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 10, ATPDIL2-2, PDI-like 2-2, PDI10, PROTEIN DISULFIDE ISOMERASE, PDIL2-2, PDI-like 2-2 |
-0.207 |
0.143 |
| 101 |
253915_at |
AT4G27280
|
[Calcium-binding EF-hand family protein] |
-0.205 |
0.164 |
| 102 |
258322_at |
AT3G22740
|
HMT3, homocysteine S-methyltransferase 3 |
-0.205 |
-0.046 |
| 103 |
253842_at |
AT4G27860
|
MEB1, MEMBRANE OF ER BODY 1 |
-0.202 |
0.093 |
| 104 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
-0.202 |
0.047 |
| 105 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
-0.201 |
0.272 |
| 106 |
261033_at |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
-0.201 |
0.370 |
| 107 |
255877_at |
AT2G40460
|
[Major facilitator superfamily protein] |
-0.198 |
-0.084 |
| 108 |
254093_at |
AT4G25110
|
AtMC2, metacaspase 2, AtMCP1c, metacaspase 1c, MC2, metacaspase 2, MCP1c, metacaspase 1c |
-0.194 |
0.270 |
| 109 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.194 |
-0.091 |
| 110 |
249932_at |
AT5G22390
|
unknown |
-0.193 |
-0.198 |
| 111 |
255254_at |
AT4G05030
|
[Copper transport protein family] |
-0.192 |
0.105 |
| 112 |
256529_at |
AT1G33260
|
[Protein kinase superfamily protein] |
-0.192 |
-0.221 |
| 113 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
-0.189 |
0.105 |
| 114 |
266799_at |
AT2G22860
|
ATPSK2, phytosulfokine 2 precursor, PSK2, phytosulfokine 2 precursor |
-0.189 |
0.202 |
| 115 |
266737_at |
AT2G47140
|
AtSDR5, SDR5, short-chain dehydrogenase reductase 5 |
-0.189 |
0.659 |
| 116 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
-0.185 |
0.062 |
| 117 |
256489_at |
AT1G31550
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.179 |
-0.146 |