|
probeID |
AGICode |
Annotation |
Log2 signal ratio
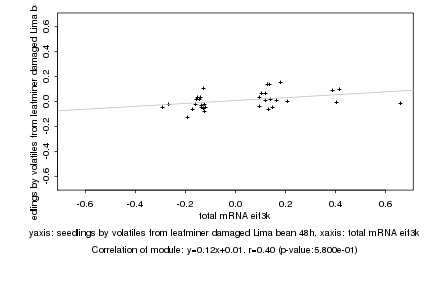
total mRNA eif3k |
Log2 signal ratio
seedlings by volatiles from leafminer damaged Lima bean 48h |
| 1 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.658 |
-0.015 |
| 2 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
0.412 |
0.100 |
| 3 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
0.403 |
-0.006 |
| 4 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
0.386 |
0.089 |
| 5 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
0.208 |
0.007 |
| 6 |
258002_at |
AT3G28930
|
AIG2, AVRRPT2-INDUCED GENE 2 |
0.179 |
0.153 |
| 7 |
245076_at |
AT2G23170
|
GH3.3 |
0.164 |
0.012 |
| 8 |
252751_at |
AT3G43430
|
[RING/U-box superfamily protein] |
0.145 |
-0.044 |
| 9 |
255108_at |
AT4G08740
|
unknown |
0.139 |
0.020 |
| 10 |
249051_at |
AT5G44390
|
[FAD-binding Berberine family protein] |
0.133 |
0.142 |
| 11 |
262277_at |
AT1G68650
|
[Uncharacterized protein family (UPF0016)] |
0.131 |
-0.056 |
| 12 |
254201_at |
AT4G24130
|
unknown |
0.125 |
0.141 |
| 13 |
257340_at |
AT2G10760
|
[copia-like retrotransposon family, has a 6.5e-285 P-value blast match to GB:CAA31653 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)] |
0.118 |
0.015 |
| 14 |
266488_at |
AT2G47670
|
PMEI6, PECTIN METHYLESTERASE INHIBITOR 6 |
0.117 |
0.068 |
| 15 |
253334_at |
AT4G33360
|
FLDH, farnesol dehydrogenase |
0.103 |
0.070 |
| 16 |
255901_at |
AT1G17890
|
GER2 |
0.094 |
0.033 |
| 17 |
266250_at |
AT2G27870
|
[similar to RNase H domain-containing protein [Arabidopsis thaliana] (TAIR:AT2G22350.1)] |
0.093 |
-0.038 |
| 18 |
261942_at |
AT1G22590
|
AGL87, AGAMOUS-like 87 |
-0.295 |
-0.044 |
| 19 |
253336_at |
AT4G33250
|
ATTIF3K1, EIF3K, eukaryotic translation initiation factor 3K, TIF3K1 |
-0.269 |
-0.023 |
| 20 |
257502_at |
AT1G78110
|
unknown |
-0.195 |
-0.121 |
| 21 |
257273_at |
AT3G28030
|
UVH3, ULTRAVIOLET HYPERSENSITIVE 3, UVR1, UV REPAIR DEFECTIVE 1 |
-0.175 |
-0.061 |
| 22 |
251981_at |
AT3G53080
|
[D-galactoside/L-rhamnose binding SUEL lectin protein] |
-0.160 |
-0.016 |
| 23 |
250696_at |
AT5G06790
|
unknown |
-0.156 |
0.021 |
| 24 |
259913_at |
AT1G72660
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.152 |
0.033 |
| 25 |
249935_at |
AT5G22420
|
FAR7, fatty acid reductase 7 |
-0.147 |
0.018 |
| 26 |
249732_at |
AT5G24420
|
PGL5, 6-phosphogluconolactonase 5 |
-0.143 |
0.034 |
| 27 |
266765_at |
AT2G46860
|
AtPPa3, pyrophosphorylase 3, PPa3, pyrophosphorylase 3 |
-0.137 |
-0.031 |
| 28 |
267037_at |
AT2G38320
|
TBL34, TRICHOME BIREFRINGENCE-LIKE 34 |
-0.136 |
-0.043 |
| 29 |
246808_at |
AT5G27110
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.129 |
0.108 |
| 30 |
258263_at |
AT3G15780
|
unknown |
-0.128 |
-0.051 |
| 31 |
254993_at |
AT4G10730
|
[Protein kinase superfamily protein] |
-0.125 |
-0.076 |
| 32 |
257337_at |
ATMG00060
|
NAD5, NADH DEHYDROGENASE SUBUNIT 5, NAD5.3, NADH DEHYDROGENASE SUBUNIT 5.3, NAD5C, NADH dehydrogenase subunit 5C |
-0.124 |
-0.022 |
| 33 |
245454_at |
AT4G16920
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.121 |
-0.041 |