|
probeID |
AGICode |
Annotation |
Log2 signal ratio
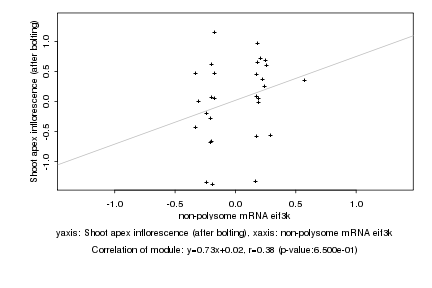
non-polysome mRNA eif3k |
Log2 signal ratio
Shoot apex inflorescence (after bolting) |
| 1 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.567 |
0.357 |
| 2 |
250503_at |
AT5G09820
|
[Plastid-lipid associated protein PAP / fibrillin family protein] |
0.289 |
-0.561 |
| 3 |
255675_at |
AT4G00480
|
ATMYC1, myc1 |
0.249 |
0.600 |
| 4 |
261080_at |
AT1G07370
|
ATPCNA1, PROLIFERATING CELLULAR NUCLEAR ANTIGEN 1, PCNA1, proliferating cellular nuclear antigen 1 |
0.242 |
0.688 |
| 5 |
246597_at |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
0.234 |
0.266 |
| 6 |
254939_at |
AT4G10800
|
unknown |
0.223 |
0.381 |
| 7 |
263535_at |
AT2G24970
|
unknown |
0.207 |
0.723 |
| 8 |
256543_at |
AT1G42480
|
unknown |
0.185 |
-0.006 |
| 9 |
248176_at |
AT5G54650
|
ATFH5, FORMIN HOMOLOGY 5, Fh5, formin homology5 |
0.183 |
0.051 |
| 10 |
247033_at |
AT5G67250
|
SKIP2, SKP1/ASK1-interacting protein 2, VFB4, VIER F-BOX PROTEINE 4 |
0.182 |
0.975 |
| 11 |
260166_at |
AT1G79840
|
GL2, GLABRA 2 |
0.178 |
0.654 |
| 12 |
253660_at |
AT4G30140
|
CDEF1, CUTICLE DESTRUCTING FACTOR 1 |
0.173 |
-0.577 |
| 13 |
259665_at |
AT1G55160
|
unknown |
0.168 |
0.458 |
| 14 |
254377_at |
AT4G21650
|
[Subtilase family protein] |
0.167 |
0.087 |
| 15 |
262671_at |
AT1G76040
|
CPK29, calcium-dependent protein kinase 29 |
0.164 |
-1.328 |
| 16 |
261942_at |
AT1G22590
|
AGL87, AGAMOUS-like 87 |
-0.336 |
-0.416 |
| 17 |
255362_at |
AT4G04000
|
[non-LTR retrotransposon family (LINE), has a 2.6e-39 P-value blast match to GB:AAB41224 ORF2 (LINE-element) (Rattus norvegicus)] |
-0.332 |
0.467 |
| 18 |
251685_at |
AT3G56430
|
unknown |
-0.310 |
0.003 |
| 19 |
257882_at |
AT3G16930
|
unknown |
-0.248 |
-0.193 |
| 20 |
255353_at |
AT4G03870
|
[Mutator-like transposase family, has a 1.1e-61 P-value blast match to O81510 /24-166 Pfam PF03108 MuDR family transposase (MuDr-element domain)] |
-0.247 |
-1.333 |
| 21 |
247264_at |
AT5G64530
|
ANAC104, Arabidopsis NAC domain containing protein 104, XND1, xylem NAC domain 1 |
-0.214 |
-0.673 |
| 22 |
255813_at |
AT1G19930
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.210 |
-0.276 |
| 23 |
261244_at |
AT1G20150
|
[Subtilisin-like serine endopeptidase family protein] |
-0.205 |
-0.659 |
| 24 |
263349_at |
AT2G13370
|
CHR5, chromatin remodeling 5 |
-0.201 |
0.627 |
| 25 |
265249_at |
AT2G01940
|
ATIDD15, ARABIDOPSIS THALIANA INDETERMINATE(ID)-DOMAIN 15, SGR5, SHOOT GRAVITROPISM 5 |
-0.200 |
0.071 |
| 26 |
252505_at |
AT3G46170
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.192 |
-1.364 |
| 27 |
253336_at |
AT4G33250
|
ATTIF3K1, EIF3K, eukaryotic translation initiation factor 3K, TIF3K1 |
-0.179 |
1.162 |
| 28 |
265991_at |
AT2G24120
|
PDE319, PIGMENT DEFECTIVE 319, SCA3, SCABRA 3 |
-0.179 |
0.475 |
| 29 |
262144_at |
AT1G52620
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.176 |
0.055 |