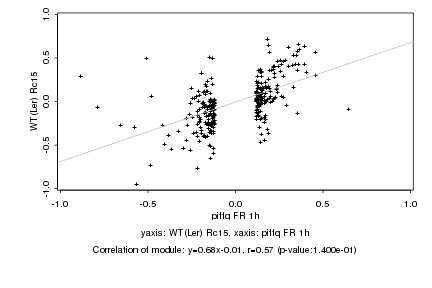
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
piffq FR 1h |
Log2 signal ratio
WT(Ler) Rc15 |
| 1 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.643 |
-0.091 |
| 2 |
246242_at |
AT4G36600
|
[Late embryogenesis abundant (LEA) protein] |
0.455 |
0.304 |
| 3 |
254440_at |
AT4G21020
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.452 |
0.566 |
| 4 |
249039_at |
AT5G44310
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.401 |
0.343 |
| 5 |
256338_at |
AT1G72100
|
[late embryogenesis abundant domain-containing protein / LEA domain-containing protein] |
0.392 |
0.429 |
| 6 |
265211_at |
AT2G36640
|
ATECP63, embryonic cell protein 63, ECP63, embryonic cell protein 63 |
0.391 |
0.639 |
| 7 |
246299_at |
AT3G51810
|
AT3, ATEM1, ARABIDOPSIS THALIANA LATE EMBRYOGENESIS ABUNDANT 1, EM1, LATE EMBRYOGENESIS ABUNDANT 1, GEA1, GUANINE NUCLEOTIDE EXCHANGE FACTOR FOR ADP RIBOSYLATION FACTORS 1 |
0.363 |
0.603 |
| 8 |
251838_at |
AT3G54940
|
[Papain family cysteine protease] |
0.358 |
0.664 |
| 9 |
258224_at |
AT3G15670
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.356 |
0.427 |
| 10 |
263753_at |
AT2G21490
|
LEA, dehydrin LEA |
0.353 |
0.579 |
| 11 |
260859_at |
AT1G43780
|
scpl44, serine carboxypeptidase-like 44 |
0.352 |
0.360 |
| 12 |
254203_at |
AT4G24150
|
AtGRF8, growth-regulating factor 8, GRF8, growth-regulating factor 8 |
0.352 |
-0.131 |
| 13 |
245335_at |
AT4G16160
|
ATOEP16-2, ATOEP16-S |
0.348 |
0.531 |
| 14 |
264494_at |
AT1G27461
|
unknown |
0.342 |
0.430 |
| 15 |
265255_at |
AT2G28420
|
GLYI8, glyoxylase I 8 |
0.327 |
0.161 |
| 16 |
258347_at |
AT3G17520
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.327 |
0.534 |
| 17 |
250624_at |
AT5G07330
|
unknown |
0.323 |
0.415 |
| 18 |
247061_at |
AT5G66780
|
unknown |
0.300 |
0.620 |
| 19 |
254095_at |
AT4G25140
|
OLE1, OLEOSIN 1, OLEO1, oleosin 1 |
0.299 |
0.409 |
| 20 |
247463_at |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
0.287 |
-0.042 |
| 21 |
259167_at |
AT3G01570
|
[Oleosin family protein] |
0.284 |
0.474 |
| 22 |
256938_at |
AT3G22500
|
ATECP31, LATE EMBRYOGENESIS ABUNDANT PROTEIN ECP31 |
0.271 |
0.047 |
| 23 |
266654_at |
AT2G25890
|
[Oleosin family protein] |
0.271 |
0.467 |
| 24 |
264888_at |
AT1G23070
|
unknown |
0.271 |
0.298 |
| 25 |
266393_at |
AT2G41260
|
ATM17, M17 |
0.262 |
0.425 |
| 26 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
0.258 |
0.058 |
| 27 |
246273_at |
AT4G36700
|
[RmlC-like cupins superfamily protein] |
0.254 |
0.471 |
| 28 |
258327_at |
AT3G22640
|
PAP85 |
0.253 |
0.348 |
| 29 |
247437_at |
AT5G62490
|
ATHVA22B, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE B, HVA22B, HVA22 homologue B |
0.245 |
0.413 |
| 30 |
249772_at |
AT5G24130
|
unknown |
0.236 |
0.188 |
| 31 |
264612_at |
AT1G04560
|
[AWPM-19-like family protein] |
0.235 |
0.461 |
| 32 |
264187_at |
AT1G54860
|
[Glycoprotein membrane precursor GPI-anchored] |
0.235 |
0.106 |
| 33 |
257059_at |
AT3G15280
|
unknown |
0.234 |
0.158 |
| 34 |
257994_at |
AT3G19920
|
unknown |
0.230 |
0.145 |
| 35 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
0.225 |
0.057 |
| 36 |
256464_at |
AT1G32560
|
AtLEA4-1, Late Embryogenesis Abundant 4-1, LEA4-1, Late Embryogenesis Abundant 4-1 |
0.223 |
0.402 |
| 37 |
256459_at |
AT1G36180
|
ACC2, acetyl-CoA carboxylase 2 |
0.223 |
0.038 |
| 38 |
266544_at |
AT2G35300
|
AtLEA4-2, late embryogenesis abundant 4-2, LEA18, late embryogenesis abundant 18, LEA4-2, late embryogenesis abundant 4-2 |
0.222 |
0.325 |
| 39 |
256352_at |
AT1G54970
|
ATPRP1, proline-rich protein 1, PRP1, proline-rich protein 1, RHS7, ROOT HAIR SPECIFIC 7 |
0.220 |
0.282 |
| 40 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
0.217 |
0.026 |
| 41 |
260917_at |
AT1G02700
|
unknown |
0.216 |
0.409 |
| 42 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.215 |
0.368 |
| 43 |
263138_at |
AT1G65090
|
unknown |
0.214 |
0.392 |
| 44 |
264347_at |
AT1G12040
|
LRX1, leucine-rich repeat/extensin 1 |
0.210 |
0.005 |
| 45 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
0.201 |
0.366 |
| 46 |
265414_at |
AT2G16660
|
[Major facilitator superfamily protein] |
0.199 |
0.124 |
| 47 |
250558_at |
AT5G07990
|
CYP75B1, CYTOCHROME P450 75B1, D501, TT7, TRANSPARENT TESTA 7 |
0.195 |
0.068 |
| 48 |
267286_at |
AT2G23640
|
RTNLB13, Reticulan like protein B13 |
0.195 |
-0.004 |
| 49 |
250418_at |
AT5G11240
|
[transducin family protein / WD-40 repeat family protein] |
0.192 |
0.160 |
| 50 |
251432_at |
AT3G59820
|
AtLETM1, LETM1, leucine zipper-EF-hand-containing transmembrane protein 1 |
0.192 |
0.035 |
| 51 |
249353_at |
AT5G40420
|
OLE2, OLEOSIN 2, OLEO2, oleosin 2 |
0.190 |
0.568 |
| 52 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
0.189 |
0.364 |
| 53 |
261305_at |
AT1G48470
|
GLN1;5, glutamine synthetase 1;5 |
0.188 |
0.255 |
| 54 |
263492_at |
AT2G42560
|
[late embryogenesis abundant domain-containing protein / LEA domain-containing protein] |
0.187 |
0.176 |
| 55 |
247582_at |
AT5G60760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.187 |
-0.357 |
| 56 |
260716_at |
AT1G48130
|
ATPER1, 1-cysteine peroxiredoxin 1, PER1, 1-cysteine peroxiredoxin 1 |
0.186 |
0.651 |
| 57 |
255893_at |
AT1G17960
|
[Threonyl-tRNA synthetase] |
0.180 |
-0.321 |
| 58 |
266511_at |
AT2G47680
|
[zinc finger (CCCH type) helicase family protein] |
0.180 |
0.032 |
| 59 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
0.178 |
0.717 |
| 60 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
0.177 |
0.150 |
| 61 |
263376_at |
AT2G20520
|
FLA6, FASCICLIN-like arabinogalactan 6 |
0.173 |
0.005 |
| 62 |
261368_at |
AT1G53070
|
[Legume lectin family protein] |
0.172 |
0.170 |
| 63 |
249784_at |
AT5G24280
|
GMI1, GAMMA-IRRADIATION AND MITOMYCIN C INDUCED 1 |
0.171 |
0.001 |
| 64 |
259042_at |
AT3G03450
|
RGL2, RGA-like 2 |
0.170 |
0.178 |
| 65 |
250214_at |
AT5G13870
|
EXGT-A4, endoxyloglucan transferase A4, XTH5, xyloglucan endotransglucosylase/hydrolase 5 |
0.170 |
-0.197 |
| 66 |
258930_at |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
0.170 |
-0.154 |
| 67 |
252137_at |
AT3G50980
|
XERO1, dehydrin xero 1 |
0.169 |
0.229 |
| 68 |
263377_at |
AT2G40220
|
ABI4, ABA INSENSITIVE 4, ATABI4, GIN6, GLUCOSE INSENSITIVE 6, ISI3, IMPAIRED SUCROSE INDUCTION 3, SAN5, SALOBRENO 5, SIS5, SUGAR-INSENSITIVE 5, SUN6, SUCROSE UNCOUPLED 6 |
0.167 |
0.097 |
| 69 |
254238_at |
AT4G23540
|
[ARM repeat superfamily protein] |
0.166 |
-0.013 |
| 70 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
0.166 |
-0.236 |
| 71 |
262347_at |
AT1G64110
|
DAA1, DUO1-activated ATPase 1 |
0.164 |
-0.443 |
| 72 |
266765_at |
AT2G46860
|
AtPPa3, pyrophosphorylase 3, PPa3, pyrophosphorylase 3 |
0.162 |
0.058 |
| 73 |
255278_at |
AT4G04940
|
[transducin family protein / WD-40 repeat family protein] |
0.162 |
0.017 |
| 74 |
248625_at |
AT5G48880
|
KAT5, 3-KETO-ACYL-COENZYME A THIOLASE 5, PKT1, PEROXISOMAL-3-KETO-ACYL-COA THIOLASE 1, PKT2, peroxisomal 3-keto-acyl-CoA thiolase 2 |
0.161 |
-0.181 |
| 75 |
256416_at |
AT3G11050
|
ATFER2, ferritin 2, FER2, ferritin 2 |
0.156 |
0.159 |
| 76 |
259411_at |
AT1G13410
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.154 |
-0.018 |
| 77 |
258965_at |
AT3G10530
|
[Transducin/WD40 repeat-like superfamily protein] |
0.152 |
0.133 |
| 78 |
263881_at |
AT2G21820
|
unknown |
0.152 |
0.169 |
| 79 |
265474_at |
AT2G15690
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.151 |
0.102 |
| 80 |
250517_at |
AT5G08260
|
scpl35, serine carboxypeptidase-like 35 |
0.150 |
0.045 |
| 81 |
245502_at |
AT4G15640
|
unknown |
0.148 |
-0.012 |
| 82 |
250180_at |
AT5G14450
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.148 |
0.336 |
| 83 |
253887_at |
AT4G27730
|
ATOPT6, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 6, OPT6, oligopeptide transporter 1 |
0.147 |
0.017 |
| 84 |
259293_at |
AT3G11580
|
[AP2/B3-like transcriptional factor family protein] |
0.147 |
-0.194 |
| 85 |
262943_at |
AT1G79470
|
[Aldolase-type TIM barrel family protein] |
0.147 |
0.055 |
| 86 |
253684_at |
AT4G29690
|
[Alkaline-phosphatase-like family protein] |
0.146 |
-0.367 |
| 87 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.146 |
0.108 |
| 88 |
251137_at |
AT5G01300
|
[PEBP (phosphatidylethanolamine-binding protein) family protein] |
0.146 |
0.350 |
| 89 |
257793_at |
AT3G26960
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.145 |
0.042 |
| 90 |
259632_at |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
0.145 |
0.207 |
| 91 |
250502_at |
AT5G09590
|
HSC70-5, HEAT SHOCK COGNATE, MTHSC70-2, mitochondrial HSO70 2 |
0.145 |
0.147 |
| 92 |
260565_at |
AT2G43800
|
[Actin-binding FH2 (formin homology 2) family protein] |
0.144 |
0.291 |
| 93 |
252238_at |
AT3G49960
|
[Peroxidase superfamily protein] |
0.143 |
0.335 |
| 94 |
259878_at |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
0.143 |
-0.113 |
| 95 |
260049_at |
AT1G29940
|
NRPA2, nuclear RNA polymerase A2 |
0.141 |
0.029 |
| 96 |
245770_at |
AT1G30240
|
[FUNCTIONS IN: binding] |
0.141 |
0.062 |
| 97 |
263515_at |
AT2G21640
|
unknown |
0.140 |
-0.461 |
| 98 |
252316_at |
AT3G48700
|
ATCXE13, carboxyesterase 13, CXE13, carboxyesterase 13 |
0.139 |
0.374 |
| 99 |
255048_at |
AT4G09600
|
GASA3, GAST1 protein homolog 3 |
0.139 |
-0.031 |
| 100 |
253011_at |
AT4G37890
|
EDA40, embryo sac development arrest 40 |
0.139 |
-0.197 |
| 101 |
256288_at |
AT3G12270
|
ATPRMT3, ARABIDOPSIS THALIANA PROTEIN ARGININE METHYLTRANSFERASE 3, PRMT3, protein arginine methyltransferase 3 |
0.139 |
0.020 |
| 102 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
0.139 |
0.151 |
| 103 |
265049_at |
AT1G52060
|
[Mannose-binding lectin superfamily protein] |
0.138 |
0.147 |
| 104 |
261023_at |
AT1G12200
|
FMO, flavin monooxygenase |
0.138 |
0.088 |
| 105 |
250683_x_at |
AT5G06640
|
EXT10, extensin 10 |
0.137 |
0.066 |
| 106 |
264471_at |
AT1G67120
|
AtMDN1, MDN1, MIDASIN 1 |
0.136 |
0.082 |
| 107 |
260955_at |
AT1G06000
|
[UDP-Glycosyltransferase superfamily protein] |
0.135 |
-0.053 |
| 108 |
245970_at |
AT5G20710
|
BGAL7, beta-galactosidase 7 |
0.133 |
-0.287 |
| 109 |
250326_at |
AT5G12080
|
ATMSL10, MSL10, mechanosensitive channel of small conductance-like 10 |
0.133 |
-0.036 |
| 110 |
259928_at |
AT1G34380
|
[5'-3' exonuclease family protein] |
0.132 |
0.078 |
| 111 |
258505_at |
AT3G06530
|
[ARM repeat superfamily protein] |
0.131 |
0.087 |
| 112 |
265345_at |
AT2G22680
|
WAVH1, WAV3 homolog 1 |
0.131 |
-0.163 |
| 113 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.130 |
0.027 |
| 114 |
262875_at |
AT1G64970
|
G-TMT, gamma-tocopherol methyltransferase, TMT1, VTE4, VITAMIN E DEFICIENT 4 |
0.130 |
0.220 |
| 115 |
251975_at |
AT3G53230
|
AtCDC48B, cell division cycle 48B |
0.130 |
-0.117 |
| 116 |
256354_at |
AT1G54870
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.129 |
0.360 |
| 117 |
248505_at |
AT5G50360
|
unknown |
0.129 |
-0.006 |
| 118 |
258765_at |
AT3G10710
|
RHS12, root hair specific 12 |
0.128 |
0.164 |
| 119 |
248984_at |
AT5G45140
|
NRPC2, nuclear RNA polymerase C2 |
0.128 |
0.022 |
| 120 |
259248_at |
AT3G07770
|
AtHsp90-6, HEAT SHOCK PROTEIN 90-6, AtHsp90.6, HEAT SHOCK PROTEIN 90.6, Hsp89.1, HEAT SHOCK PROTEIN 89.1 |
0.128 |
0.031 |
| 121 |
266510_at |
AT2G47990
|
EDA13, EMBRYO SAC DEVELOPMENT ARREST 13, EDA19, EMBRYO SAC DEVELOPMENT ARREST 19, SWA1, SLOW WALKER1 |
0.128 |
0.070 |
| 122 |
248246_at |
AT5G53200
|
TRY, TRIPTYCHON |
0.128 |
-0.151 |
| 123 |
245163_at |
AT2G33230
|
YUC7, YUCCA 7 |
0.127 |
-0.015 |
| 124 |
260824_at |
AT1G06720
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.126 |
-0.087 |
| 125 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
0.125 |
0.114 |
| 126 |
260876_at |
AT1G21460
|
AtSWEET1, SWEET1 |
0.125 |
0.213 |
| 127 |
267044_at |
AT2G34357
|
[ARM repeat superfamily protein] |
0.125 |
0.107 |
| 128 |
247268_at |
AT5G64080
|
AtXYP1, XYP1, xylogen protein 1 |
0.125 |
0.290 |
| 129 |
247914_at |
AT5G57540
|
AtXTH13, XTH13, xyloglucan endotransglucosylase/hydrolase 13 |
0.125 |
0.188 |
| 130 |
253782_at |
AT4G28590
|
AtECB1, ECB1, early chloroplast biogenesis 1, MRL7, Mesophyll-cell RNAi Library line 7, SVR4, SUPPRESSOR OF VARIEGATION 4 |
0.123 |
0.103 |
| 131 |
249354_at |
AT5G40480
|
EMB3012, embryo defective 3012 |
0.123 |
-0.037 |
| 132 |
259018_at |
AT3G07390
|
AIR12, Auxin-Induced in Root cultures 12 |
0.123 |
-0.049 |
| 133 |
257608_at |
AT3G13860
|
HSP60-3A, heat shock protein 60-3A |
0.123 |
0.036 |
| 134 |
247989_at |
AT5G56350
|
[Pyruvate kinase family protein] |
0.122 |
-0.065 |
| 135 |
263962_at |
AT2G36350
|
[Protein kinase superfamily protein] |
0.122 |
-0.083 |
| 136 |
248590_at |
AT5G49660
|
XIP1, XYLEM INTERMIXED WITH PHLOEM 1 |
0.122 |
0.001 |
| 137 |
260331_at |
AT1G80270
|
PPR596, PENTATRICOPEPTIDE REPEAT 596 |
0.122 |
0.048 |
| 138 |
254097_at |
AT4G25160
|
[U-box domain-containing protein kinase family protein] |
0.122 |
-0.137 |
| 139 |
246138_at |
AT5G19870
|
unknown |
0.122 |
-0.150 |
| 140 |
249830_at |
AT5G23300
|
PYRD, pyrimidine d |
0.121 |
0.077 |
| 141 |
266462_at |
AT2G47770
|
ATTSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related, TSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related |
0.121 |
0.157 |
| 142 |
248709_at |
AT5G48470
|
PRDA1, PEP-Related Development Arrested 1 |
0.120 |
0.010 |
| 143 |
258794_at |
AT3G04710
|
TPR10, tetratricopeptide repeat 10 |
0.120 |
0.008 |
| 144 |
264723_at |
AT1G22960
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.120 |
0.032 |
| 145 |
253600_at |
AT4G30810
|
scpl29, serine carboxypeptidase-like 29 |
0.120 |
0.046 |
| 146 |
252377_at |
AT3G47960
|
AtNPF2.10, GTR1, GLUCOSINOLATE TRANSPORTER-1, NPF2.10, NRT1/ PTR family 2.10 |
0.120 |
0.076 |
| 147 |
260023_at |
AT1G30040
|
ATGA2OX2, gibberellin 2-oxidase, GA2OX2, gibberellin 2-oxidase, GA2OX2, GIBBERELLIN 2-OXIDASE 2 |
0.119 |
-0.171 |
| 148 |
249456_at |
AT5G39410
|
[Saccharopine dehydrogenase ] |
0.118 |
-0.118 |
| 149 |
262819_at |
AT1G11600
|
CYP77B1, cytochrome P450, family 77, subfamily B, polypeptide 1 |
0.118 |
-0.204 |
| 150 |
251583_at |
AT3G58590
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.118 |
0.016 |
| 151 |
256966_at |
AT3G13400
|
sks13, SKU5 similar 13 |
0.117 |
-0.095 |
| 152 |
261691_at |
AT1G50060
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.117 |
0.230 |
| 153 |
248307_at |
AT5G52850
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.117 |
0.112 |
| 154 |
248961_at |
AT5G45650
|
[subtilase family protein] |
0.117 |
0.094 |
| 155 |
265930_at |
AT2G18510
|
emb2444, embryo defective 2444 |
0.117 |
0.106 |
| 156 |
261762_at |
AT1G15510
|
ATECB2, ARABIDOPSIS EARLY CHLOROPLAST BIOGENESIS2, ECB2, EARLY CHLOROPLAST BIOGENESIS2, VAC1, VANILLA CREAM 1 |
0.115 |
0.117 |
| 157 |
261330_at |
AT1G44900
|
ATMCM2, MCM2, MINICHROMOSOME MAINTENANCE 2 |
0.115 |
0.029 |
| 158 |
251497_at |
AT3G59060
|
PIF5, PHYTOCHROME-INTERACTING FACTOR 5, PIL6, phytochrome interacting factor 3-like 6 |
-0.888 |
0.293 |
| 159 |
264510_at |
AT1G09530
|
PAP3, PHYTOCHROME-ASSOCIATED PROTEIN 3, PIF3, phytochrome interacting factor 3, POC1, PHOTOCURRENT 1 |
-0.793 |
-0.061 |
| 160 |
259785_at |
AT1G29490
|
SAUR68, SMALL AUXIN UPREGULATED 68 |
-0.657 |
-0.266 |
| 161 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
-0.582 |
-0.295 |
| 162 |
254685_at |
AT4G13790
|
SAUR25, SMALL AUXIN UPREGULATED RNA 25 |
-0.570 |
-0.941 |
| 163 |
265248_at |
AT2G43010
|
AtPIF4, PIF4, phytochrome interacting factor 4, SRL2 |
-0.512 |
0.504 |
| 164 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
-0.489 |
-0.724 |
| 165 |
265584_at |
AT2G20180
|
PIF1, PHY-INTERACTING FACTOR 1, PIL5, phytochrome interacting factor 3-like 5 |
-0.480 |
0.062 |
| 166 |
252282_at |
AT3G49360
|
PGL2, 6-phosphogluconolactonase 2 |
-0.422 |
-0.270 |
| 167 |
245276_at |
AT4G16780
|
ATHB-2, homeobox protein 2, ATHB2, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2, HAT4, HB-2, homeobox protein 2 |
-0.410 |
-0.489 |
| 168 |
266814_at |
AT2G44910
|
ATHB-4, ARABIDOPSIS THALIANA HOMEOBOX-LEUCINE ZIPPER PROTEIN 4, ATHB4, homeobox-leucine zipper protein 4, HB4, homeobox-leucine zipper protein 4 |
-0.388 |
-0.389 |
| 169 |
253155_at |
AT4G35720
|
unknown |
-0.369 |
-0.541 |
| 170 |
259834_at |
AT1G69570
|
[Dof-type zinc finger DNA-binding family protein] |
-0.328 |
-0.339 |
| 171 |
261109_at |
AT1G75450
|
ATCKX5, ARABIDOPSIS THALIANA CYTOKININ OXIDASE 5, ATCKX6, CYTOKININ OXIDASE 6, CKX5, cytokinin oxidase 5 |
-0.299 |
-0.531 |
| 172 |
260135_at |
AT1G66400
|
CML23, calmodulin like 23 |
-0.285 |
-0.194 |
| 173 |
256743_at |
AT3G29370
|
P1R3, P1R3 |
-0.282 |
-0.446 |
| 174 |
251774_at |
AT3G55840
|
[Hs1pro-1 protein] |
-0.279 |
-0.274 |
| 175 |
261050_at |
AT1G01260
|
JAM2, Jasmonate Associated MYC2 LIKE 2 |
-0.267 |
-0.143 |
| 176 |
251870_at |
AT3G54510
|
[Early-responsive to dehydration stress protein (ERD4)] |
-0.262 |
-0.554 |
| 177 |
257650_at |
AT3G16800
|
[Protein phosphatase 2C family protein] |
-0.260 |
-0.020 |
| 178 |
250155_at |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
-0.252 |
0.155 |
| 179 |
260371_at |
AT1G69690
|
AtTCP15, TCP15, TEOSINTE BRANCHED1/CYCLOIDEA/PCF 15 |
-0.248 |
-0.172 |
| 180 |
261292_at |
AT1G36940
|
unknown |
-0.246 |
0.040 |
| 181 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.240 |
-0.360 |
| 182 |
250971_at |
AT5G02810
|
APRR7, PRR7, pseudo-response regulator 7 |
-0.234 |
0.043 |
| 183 |
257929_at |
AT3G16980
|
NRPB9A, NRPD9A, NRPE9A |
-0.227 |
0.060 |
| 184 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-0.224 |
-0.348 |
| 185 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
-0.220 |
-0.104 |
| 186 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
-0.219 |
-0.150 |
| 187 |
258395_at |
AT3G15500
|
ANAC055, NAC domain containing protein 55, ATNAC3, NAC domain containing protein 3, NAC055, NAC domain containing protein 55, NAC3, NAC domain containing protein 3 |
-0.219 |
-0.255 |
| 188 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-0.218 |
-0.392 |
| 189 |
251012_at |
AT5G02580
|
unknown |
-0.217 |
-0.766 |
| 190 |
254716_at |
AT4G13560
|
UNE15, unfertilized embryo sac 15 |
-0.216 |
0.120 |
| 191 |
247128_at |
AT5G66110
|
HIPP27, heavy metal associated isoprenylated plant protein 27 |
-0.212 |
0.011 |
| 192 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
-0.211 |
0.073 |
| 193 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
-0.211 |
-0.235 |
| 194 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
-0.210 |
-0.082 |
| 195 |
264697_at |
AT1G70210
|
ATCYCD1;1, CYCD1;1, CYCLIN D1;1 |
-0.209 |
-0.173 |
| 196 |
245439_at |
AT4G16670
|
unknown |
-0.209 |
-0.449 |
| 197 |
248208_at |
AT5G53980
|
ATHB52, homeobox protein 52, HB52, homeobox protein 52 |
-0.209 |
-0.224 |
| 198 |
245861_at |
AT5G28300
|
AtGT2L, GT2L, GT-2Like protein |
-0.208 |
-0.178 |
| 199 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.202 |
-0.195 |
| 200 |
248160_at |
AT5G54470
|
BBX29, B-box domain protein 29 |
-0.202 |
-0.298 |
| 201 |
254634_at |
AT4G18650
|
[transcription factor-related] |
-0.197 |
0.322 |
| 202 |
254193_at |
AT4G23870
|
unknown |
-0.197 |
-0.322 |
| 203 |
254305_at |
AT4G22200
|
AKT2, AKT2/3, potassium transport 2/3, AKT3, KT2/3, potassium transport 2/3 |
-0.196 |
0.325 |
| 204 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
-0.194 |
-0.356 |
| 205 |
266976_at |
AT2G39410
|
[alpha/beta-Hydrolases superfamily protein] |
-0.192 |
-0.064 |
| 206 |
262549_at |
AT1G31290
|
AGO3, ARGONAUTE 3 |
-0.192 |
-0.013 |
| 207 |
260744_at |
AT1G15010
|
unknown |
-0.187 |
0.111 |
| 208 |
261611_at |
AT1G49730
|
[Protein kinase superfamily protein] |
-0.187 |
-0.002 |
| 209 |
252129_at |
AT3G50890
|
AtHB28, homeobox protein 28, HB28, homeobox protein 28, ZHD7, ZINC FINGER HOMEODOMAIN 7 |
-0.186 |
-0.399 |
| 210 |
254178_at |
AT4G23880
|
unknown |
-0.183 |
-0.025 |
| 211 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
-0.182 |
-0.381 |
| 212 |
247880_at |
AT5G57780
|
P1R1, P1R1 |
-0.178 |
-0.064 |
| 213 |
267613_at |
AT2G26700
|
PID2, PINOID2 |
-0.178 |
0.125 |
| 214 |
266164_at |
AT2G28050
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.178 |
-0.129 |
| 215 |
264223_s_at |
AT3G16030
|
CES101, CALLUS EXPRESSION OF RBCS 101, RFO3, RESISTANCE TO FUSARIUM OXYSPORUM 3 |
-0.178 |
-0.137 |
| 216 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
-0.177 |
-0.027 |
| 217 |
253913_at |
AT4G27370
|
ATVIIIB, MYOSIN VIII B, VIIIB |
-0.176 |
0.090 |
| 218 |
247927_at |
AT5G57310
|
unknown |
-0.176 |
-0.048 |
| 219 |
255723_at |
AT3G29575
|
AFP3, ABI five binding protein 3 |
-0.175 |
-0.236 |
| 220 |
250158_at |
AT5G15190
|
unknown |
-0.173 |
-0.119 |
| 221 |
247878_at |
AT5G57760
|
unknown |
-0.173 |
-0.403 |
| 222 |
264815_at |
AT1G03620
|
[ELMO/CED-12 family protein] |
-0.172 |
0.200 |
| 223 |
248509_at |
AT5G50335
|
unknown |
-0.171 |
-0.164 |
| 224 |
255285_at |
AT4G04630
|
unknown |
-0.170 |
0.177 |
| 225 |
266021_at |
AT2G05910
|
unknown |
-0.167 |
-0.171 |
| 226 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.167 |
-0.255 |
| 227 |
259645_at |
AT1G69010
|
BIM2, BES1-interacting Myc-like protein 2 |
-0.166 |
-0.049 |
| 228 |
257863_at |
AT3G17730
|
anac057, NAC domain containing protein 57, NAC057, NAC domain containing protein 57 |
-0.166 |
0.123 |
| 229 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.164 |
-0.255 |
| 230 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.163 |
-0.401 |
| 231 |
250327_at |
AT5G12050
|
unknown |
-0.162 |
0.237 |
| 232 |
250975_at |
AT5G03050
|
unknown |
-0.162 |
0.118 |
| 233 |
264512_at |
AT1G09575
|
unknown |
-0.161 |
-0.081 |
| 234 |
251601_at |
AT3G57800
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.161 |
-0.061 |
| 235 |
260004_at |
AT1G67860
|
unknown |
-0.161 |
-0.251 |
| 236 |
248197_at |
AT5G54190
|
PORA, protochlorophyllide oxidoreductase A |
-0.157 |
-0.256 |
| 237 |
254110_at |
AT4G25260
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.157 |
-0.307 |
| 238 |
253617_at |
AT4G30410
|
[sequence-specific DNA binding transcription factors] |
-0.155 |
-0.232 |
| 239 |
256066_at |
AT1G06980
|
unknown |
-0.154 |
-0.204 |
| 240 |
249807_at |
AT5G23870
|
[Pectinacetylesterase family protein] |
-0.154 |
-0.149 |
| 241 |
245777_at |
AT1G73540
|
atnudt21, nudix hydrolase homolog 21, NUDT21, nudix hydrolase homolog 21 |
-0.153 |
0.007 |
| 242 |
265276_at |
AT2G28400
|
unknown |
-0.153 |
-0.154 |
| 243 |
264774_at |
AT1G22890
|
unknown |
-0.151 |
0.511 |
| 244 |
245440_at |
AT4G16680
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.150 |
-0.045 |
| 245 |
255854_at |
AT1G67050
|
unknown |
-0.150 |
-0.056 |
| 246 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
-0.149 |
-0.345 |
| 247 |
265245_at |
AT2G43060
|
AtIBH1, IBH1, ILI1 binding bHLH 1 |
-0.149 |
-0.497 |
| 248 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
-0.148 |
-0.653 |
| 249 |
260527_at |
AT2G47270
|
UPB1, UPBEAT1 |
-0.148 |
-0.332 |
| 250 |
266756_at |
AT2G46950
|
CYP709B2, cytochrome P450, family 709, subfamily B, polypeptide 2 |
-0.148 |
-0.009 |
| 251 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
-0.146 |
-0.267 |
| 252 |
254901_at |
AT4G11530
|
CRK34, cysteine-rich RLK (RECEPTOR-like protein kinase) 34 |
-0.146 |
0.011 |
| 253 |
258399_at |
AT3G15540
|
IAA19, indole-3-acetic acid inducible 19, MSG2, MASSUGU 2 |
-0.145 |
-0.516 |
| 254 |
245734_at |
AT1G73480
|
[alpha/beta-Hydrolases superfamily protein] |
-0.145 |
0.096 |
| 255 |
251374_at |
AT3G60390
|
HAT3, homeobox-leucine zipper protein 3 |
-0.144 |
-0.130 |
| 256 |
255160_at |
AT4G07820
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.144 |
0.002 |
| 257 |
256257_at |
AT3G11240
|
ATATE2, ATE2, arginine-tRNA protein transferase 2 |
-0.143 |
0.024 |
| 258 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-0.143 |
0.014 |
| 259 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
-0.143 |
-0.106 |
| 260 |
251245_at |
AT3G62090
|
PIF6, PHYTOCHROME-INTERACTING FACTOR 6, PIL2, phytochrome interacting factor 3-like 2 |
-0.142 |
-0.359 |
| 261 |
253332_at |
AT4G33420
|
[Peroxidase superfamily protein] |
-0.141 |
-0.154 |
| 262 |
251039_at |
AT5G02020
|
SIS, Salt Induced Serine rich |
-0.141 |
-0.075 |
| 263 |
256740_at |
AT3G29330
|
unknown |
-0.141 |
0.013 |
| 264 |
253207_at |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
-0.141 |
-0.255 |
| 265 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
-0.141 |
0.266 |
| 266 |
254681_at |
AT4G18140
|
SSP4b, SCP1-like small phosphatase 4b |
-0.139 |
-0.083 |
| 267 |
266799_at |
AT2G22860
|
ATPSK2, phytosulfokine 2 precursor, PSK2, phytosulfokine 2 precursor |
-0.138 |
-0.282 |
| 268 |
258362_at |
AT3G14280
|
unknown |
-0.138 |
-0.334 |
| 269 |
252133_at |
AT3G50900
|
unknown |
-0.138 |
-0.232 |
| 270 |
251771_at |
AT3G56000
|
ATCSLA14, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE A14, CSLA14, cellulose synthase like A14 |
-0.137 |
-0.249 |
| 271 |
251430_at |
AT3G60110
|
[DNA-binding bromodomain-containing protein] |
-0.137 |
-0.050 |
| 272 |
248432_at |
AT5G51390
|
unknown |
-0.136 |
-0.194 |
| 273 |
247047_at |
AT5G66650
|
unknown |
-0.136 |
-0.256 |
| 274 |
261456_at |
AT1G21050
|
unknown |
-0.135 |
-0.038 |
| 275 |
266555_at |
AT2G46270
|
GBF3, G-box binding factor 3 |
-0.135 |
-0.047 |
| 276 |
249259_at |
AT5G41660
|
unknown |
-0.133 |
-0.065 |
| 277 |
247455_at |
AT5G62470
|
ATMYB96, MYB DOMAIN PROTEIN 96, MYB96, myb domain protein 96, MYBCOV1 |
-0.132 |
-0.036 |
| 278 |
265323_at |
AT2G18260
|
ATSYP112, SYP112, syntaxin of plants 112 |
-0.132 |
0.205 |
| 279 |
259093_at |
AT3G04860
|
unknown |
-0.132 |
-0.029 |
| 280 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
-0.132 |
0.498 |
| 281 |
251925_at |
AT3G54000
|
unknown |
-0.131 |
-0.137 |
| 282 |
261984_at |
AT1G33760
|
[Integrase-type DNA-binding superfamily protein] |
-0.131 |
0.015 |
| 283 |
247352_at |
AT5G63650
|
SNRK2-5, SNRK2.5, SNF1-related protein kinase 2.5, SRK2H, SNF1-RELATED PROTEIN KINASE 2H |
-0.130 |
-0.531 |
| 284 |
262301_at |
AT1G70880
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.130 |
-0.054 |
| 285 |
250425_at |
AT5G10530
|
LecRK-IX.1, L-type lectin receptor kinase IX.1 |
-0.130 |
-0.364 |
| 286 |
252970_at |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
-0.130 |
-0.336 |
| 287 |
255753_at |
AT1G18570
|
AtMYB51, myb domain protein 51, BW51A, BW51B, HIG1, HIGH INDOLIC GLUCOSINOLATE 1, MYB51, myb domain protein 51 |
-0.129 |
-0.169 |
| 288 |
258385_at |
AT3G15510
|
ANAC056, Arabidopsis NAC domain containing protein 56, ATNAC2, NAC domain containing protein 2, NAC2, NAC domain containing protein 2, NARS1, NAC-REGULATED SEED MORPHOLOGY 1 |
-0.128 |
-0.248 |
| 289 |
255511_at |
AT4G02075
|
PIT1, pitchoun 1 |
-0.128 |
-0.233 |
| 290 |
264395_at |
AT1G12070
|
[Immunoglobulin E-set superfamily protein] |
-0.128 |
-0.223 |
| 291 |
253813_at |
AT4G28150
|
unknown |
-0.127 |
-0.083 |
| 292 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
-0.127 |
-0.177 |
| 293 |
248110_at |
AT5G55320
|
[MBOAT (membrane bound O-acyl transferase) family protein] |
-0.127 |
-0.133 |
| 294 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
-0.127 |
-0.048 |
| 295 |
245336_at |
AT4G16515
|
CLEL 6, CLE-like 6, GLV1, GOLVEN 1, RGF6, root meristem growth factor 6 |
-0.126 |
-0.239 |
| 296 |
250748_at |
AT5G05710
|
[Pleckstrin homology (PH) domain superfamily protein] |
-0.126 |
-0.028 |
| 297 |
265588_at |
AT2G19970
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.126 |
-0.293 |
| 298 |
247074_at |
AT5G66590
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.126 |
-0.587 |
| 299 |
254362_at |
AT4G22160
|
unknown |
-0.126 |
-0.098 |
| 300 |
250665_at |
AT5G06980
|
LNK4, night light-inducible and clock-regulated 4 |
-0.126 |
0.002 |
| 301 |
255642_at |
AT4G00820
|
iqd17, IQ-domain 17 |
-0.125 |
-0.211 |
| 302 |
252557_at |
AT3G45960
|
ATEXLA3, expansin-like A3, ATEXPL3, ATHEXP BETA 2.3, EXLA3, expansin-like A3, EXPL3 |
-0.125 |
-0.241 |
| 303 |
259839_at |
AT1G52190
|
AtNPF1.2, NPF1.2, NRT1/ PTR family 1.2, NRT1.11, nitrate transporter 1.11 |
-0.125 |
-0.020 |
| 304 |
261451_at |
AT1G21060
|
unknown |
-0.125 |
0.020 |
| 305 |
266983_at |
AT2G39400
|
[alpha/beta-Hydrolases superfamily protein] |
-0.124 |
-0.086 |
| 306 |
260819_at |
AT1G06850
|
AtbZIP52, basic leucine-zipper 52, bZIP52, basic leucine-zipper 52 |
-0.124 |
-0.256 |
| 307 |
258275_at |
AT3G15760
|
unknown |
-0.124 |
-0.148 |
| 308 |
266613_at |
AT2G14900
|
[Gibberellin-regulated family protein] |
-0.123 |
-0.207 |
| 309 |
257569_at |
AT3G26350
|
[LOCATED IN: chloroplast] |
-0.122 |
0.030 |
| 310 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
-0.122 |
-0.004 |
| 311 |
262850_at |
AT1G14920
|
GAI, GIBBERELLIC ACID INSENSITIVE, RGA2, RESTORATION ON GROWTH ON AMMONIA 2 |
-0.122 |
-0.016 |
| 312 |
251903_at |
AT3G54120
|
[Reticulon family protein] |
-0.122 |
-0.156 |
| 313 |
256377_at |
AT1G66650
|
[Protein with RING/U-box and TRAF-like domains] |
-0.122 |
-0.036 |