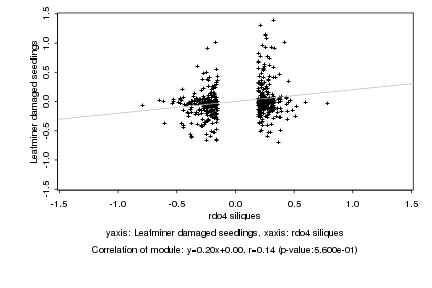
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
rdo4 siliques |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
260459_at |
AT1G68240
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.782 |
-0.032 |
| 2 |
246055_at |
AT5G08380
|
AGAL1, alpha-galactosidase 1, AtAGAL1, alpha-galactosidase 1 |
0.596 |
-0.002 |
| 3 |
249742_at |
AT5G24490
|
[30S ribosomal protein, putative] |
0.517 |
-0.080 |
| 4 |
254059_at |
AT4G25200
|
ATHSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6, HSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6 |
0.509 |
-0.256 |
| 5 |
257891_at |
AT3G17170
|
RFC3, REGULATOR OF FATTY-ACID COMPOSITION 3 |
0.481 |
-0.144 |
| 6 |
256045_at |
AT1G07150
|
MAPKKK13, mitogen-activated protein kinase kinase kinase 13 |
0.461 |
0.024 |
| 7 |
258339_at |
AT3G16120
|
[Dynein light chain type 1 family protein] |
0.449 |
-0.013 |
| 8 |
247450_at |
AT5G62350
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.447 |
0.350 |
| 9 |
264176_at |
AT1G02110
|
unknown |
0.443 |
-0.304 |
| 10 |
257306_at |
AT3G30200
|
[Plant transposase (Ptta/En/Spm family)] |
0.439 |
-0.155 |
| 11 |
246246_at |
AT4G37170
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.435 |
-0.038 |
| 12 |
265992_at |
AT2G24130
|
[Leucine-rich receptor-like protein kinase family protein] |
0.428 |
0.053 |
| 13 |
254934_at |
AT4G11140
|
CRF1, cytokinin response factor 1 |
0.428 |
0.080 |
| 14 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
0.411 |
1.022 |
| 15 |
250211_at |
AT5G13880
|
unknown |
0.392 |
0.091 |
| 16 |
248614_at |
AT5G49560
|
[Putative methyltransferase family protein] |
0.387 |
-0.168 |
| 17 |
265584_at |
AT2G20180
|
PIF1, PHY-INTERACTING FACTOR 1, PIL5, phytochrome interacting factor 3-like 5 |
0.383 |
-0.488 |
| 18 |
260978_at |
AT1G53540
|
[HSP20-like chaperones superfamily protein] |
0.383 |
-0.272 |
| 19 |
245103_at |
AT2G41590
|
unknown |
0.381 |
0.049 |
| 20 |
255811_at |
AT4G10250
|
ATHSP22.0 |
0.380 |
0.006 |
| 21 |
259444_at |
AT1G02370
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.378 |
-0.118 |
| 22 |
250517_at |
AT5G08260
|
scpl35, serine carboxypeptidase-like 35 |
0.375 |
-0.258 |
| 23 |
264390_at |
AT1G11950
|
[Transcription factor jumonji (jmjC) domain-containing protein] |
0.374 |
0.058 |
| 24 |
259947_at |
AT1G71530
|
[Protein kinase superfamily protein] |
0.374 |
0.083 |
| 25 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
0.368 |
0.473 |
| 26 |
265147_at |
AT1G51380
|
[DEA(D/H)-box RNA helicase family protein] |
0.365 |
-0.008 |
| 27 |
264554_at |
AT1G09510
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.359 |
0.041 |
| 28 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
0.359 |
-0.687 |
| 29 |
264167_at |
AT1G02060
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.355 |
-0.111 |
| 30 |
254263_at |
AT4G23493
|
unknown |
0.354 |
-0.172 |
| 31 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
0.344 |
-0.256 |
| 32 |
251304_at |
AT3G61990
|
OMTF3, O-MTase family 3 protein |
0.336 |
0.425 |
| 33 |
253083_at |
AT4G36250
|
ALDH3F1, aldehyde dehydrogenase 3F1 |
0.332 |
-0.098 |
| 34 |
248021_at |
AT5G56500
|
Cpn60beta3, chaperonin-60beta3 |
0.330 |
-0.123 |
| 35 |
262838_at |
AT1G14960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.325 |
-0.003 |
| 36 |
247799_at |
AT5G58840
|
[Subtilase family protein] |
0.325 |
0.019 |
| 37 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
0.325 |
0.915 |
| 38 |
264280_at |
AT1G61820
|
BGLU46, beta glucosidase 46 |
0.323 |
0.416 |
| 39 |
259415_at |
AT1G02330
|
unknown |
0.322 |
0.092 |
| 40 |
250087_at |
AT5G17270
|
[Protein prenylyltransferase superfamily protein] |
0.322 |
-0.124 |
| 41 |
247196_at |
AT5G65510
|
AIL7, AINTEGUMENTA-like 7, PLT7, PLETHORA 7 |
0.322 |
-0.001 |
| 42 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
0.320 |
1.403 |
| 43 |
250746_at |
AT5G05880
|
[UDP-Glycosyltransferase superfamily protein] |
0.320 |
-0.013 |
| 44 |
261586_at |
AT1G01640
|
[BTB/POZ domain-containing protein] |
0.320 |
0.017 |
| 45 |
254043_at |
AT4G25990
|
CIL |
0.318 |
-0.282 |
| 46 |
245032_at |
AT2G26630
|
[transposase IS4 family protein, contains Pfam profile: PF01609 transposase DDE domain] |
0.317 |
-0.059 |
| 47 |
261923_at |
AT1G22380
|
AtUGT85A3, UDP-glucosyl transferase 85A3, UGT85A3, UDP-glucosyl transferase 85A3 |
0.317 |
0.055 |
| 48 |
263406_at |
AT2G04160
|
AIR3, AUXIN-INDUCED IN ROOT CULTURES 3 |
0.316 |
-0.026 |
| 49 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
0.316 |
0.598 |
| 50 |
262148_at |
AT1G52560
|
[HSP20-like chaperones superfamily protein] |
0.315 |
-0.005 |
| 51 |
249836_at |
AT5G23420
|
HMGB6, high-mobility group box 6 |
0.313 |
-0.147 |
| 52 |
261655_at |
AT1G01940
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
0.311 |
-0.058 |
| 53 |
253780_at |
AT4G28400
|
[Protein phosphatase 2C family protein] |
0.311 |
0.127 |
| 54 |
253092_at |
AT4G37380
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.309 |
-0.166 |
| 55 |
257112_at |
AT3G20120
|
CYP705A21, cytochrome P450, family 705, subfamily A, polypeptide 21 |
0.308 |
-0.076 |
| 56 |
257830_at |
AT3G26690
|
ATNUDT13, ARABIDOPSIS THALIANA NUDIX HYDROLASE HOMOLOG 13, ATNUDX13, nudix hydrolase homolog 13, NUDX13, nudix hydrolase homolog 13 |
0.307 |
-0.027 |
| 57 |
264175_at |
AT1G02050
|
LAP6, LESS ADHESIVE POLLEN 6, PSKA, polyketide synthase A |
0.305 |
-0.097 |
| 58 |
262322_at |
AT1G27590
|
unknown |
0.304 |
-0.109 |
| 59 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
0.302 |
-0.379 |
| 60 |
255732_at |
AT1G25450
|
CER60, ECERIFERUM 60, KCS5, 3-ketoacyl-CoA synthase 5 |
0.301 |
0.046 |
| 61 |
259445_at |
AT1G02400
|
ATGA2OX4, Arabidopsis thaliana gibberellin 2-oxidase 4, ATGA2OX6, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 6, DTA1, DOWNSTREAM TARGET OF AGL15 1, GA2OX6, gibberellin 2-oxidase 6 |
0.301 |
0.932 |
| 62 |
263679_at |
AT1G59990
|
EMB3108, EMBRYO DEFECTIVE 3108, HS3, HEAVY SEED 3, RH22, RNA helicase 22 |
0.299 |
-0.076 |
| 63 |
250196_at |
AT5G14580
|
[polyribonucleotide nucleotidyltransferase, putative] |
0.298 |
-0.116 |
| 64 |
264806_at |
AT1G08610
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.298 |
-0.170 |
| 65 |
251751_at |
AT3G55720
|
unknown |
0.298 |
0.034 |
| 66 |
250766_at |
AT5G05550
|
[sequence-specific DNA binding transcription factors] |
0.296 |
-0.099 |
| 67 |
256073_at |
AT1G18100
|
E12A11, MFT, MOTHER OF FT AND TFL1 |
0.295 |
0.003 |
| 68 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
0.293 |
-0.517 |
| 69 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
0.290 |
0.748 |
| 70 |
245555_at |
AT4G15390
|
[HXXXD-type acyl-transferase family protein] |
0.290 |
0.009 |
| 71 |
253975_at |
AT4G26600
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.286 |
-0.165 |
| 72 |
260528_at |
AT2G47260
|
ATWRKY23, WRKY DNA-BINDING PROTEIN 23, WRKY23, WRKY DNA-binding protein 23 |
0.286 |
0.082 |
| 73 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
0.286 |
0.444 |
| 74 |
253392_at |
AT4G32650
|
ATKC1, ARABIDOPSIS THALIANA K+ RECTIFYING CHANNEL 1, AtLKT1, A. thaliana low-K+-tolerant 1, KAT3, potassium channel in Arabidopsis thaliana 3, KC1 |
0.285 |
0.294 |
| 75 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.284 |
0.621 |
| 76 |
260585_at |
AT2G43650
|
EMB2777, EMBRYO DEFECTIVE 2777 |
0.283 |
-0.184 |
| 77 |
267199_at |
AT2G30990
|
unknown |
0.281 |
0.165 |
| 78 |
253566_at |
AT4G31210
|
[DNA topoisomerase, type IA, core] |
0.281 |
-0.176 |
| 79 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
0.279 |
-0.516 |
| 80 |
264142_at |
AT1G78930
|
[Mitochondrial transcription termination factor family protein] |
0.278 |
-0.221 |
| 81 |
258631_at |
AT3G07970
|
QRT2, QUARTET 2 |
0.277 |
0.046 |
| 82 |
246254_at |
AT4G36450
|
ATMPK14, mitogen-activated protein kinase 14, MPK14, MPK14, mitogen-activated protein kinase 14 |
0.277 |
0.037 |
| 83 |
256371_at |
AT1G66770
|
AtSWEET6, SWEET6 |
0.277 |
-0.045 |
| 84 |
251985_at |
AT3G53220
|
[Thioredoxin superfamily protein] |
0.277 |
-0.013 |
| 85 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
0.276 |
0.384 |
| 86 |
257678_at |
AT3G20420
|
ATRTL2, RNASEIII-LIKE 2, RTL2, RNAse THREE-like protein 2 |
0.276 |
-0.097 |
| 87 |
249443_at |
AT5G39600
|
unknown |
0.275 |
0.003 |
| 88 |
267276_at |
AT2G30130
|
ASL5, LBD12, PCK1, PEACOCK 1 |
0.274 |
-0.019 |
| 89 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.274 |
0.361 |
| 90 |
251155_at |
AT3G63160
|
OEP6, outer envelope protein 6 |
0.273 |
-0.406 |
| 91 |
257717_at |
AT3G18390
|
EMB1865, embryo defective 1865 |
0.272 |
-0.231 |
| 92 |
256284_at |
AT3G12530
|
PSF2 |
0.271 |
0.044 |
| 93 |
258397_at |
AT3G15357
|
unknown |
0.270 |
0.137 |
| 94 |
250762_at |
AT5G05990
|
[Mitochondrial glycoprotein family protein] |
0.269 |
-0.117 |
| 95 |
250646_at |
AT5G06720
|
ATPA2, peroxidase 2, AtPRX53, PA2, peroxidase 2, PRX53, peroxidase 53 |
0.269 |
0.230 |
| 96 |
247192_at |
AT5G65360
|
H3.1, histone 3.1 |
0.269 |
-0.039 |
| 97 |
254839_at |
AT4G12400
|
Hop3, Hop3 |
0.267 |
-0.588 |
| 98 |
257026_at |
AT3G19200
|
unknown |
0.267 |
0.013 |
| 99 |
248333_at |
AT5G52390
|
[PAR1 protein] |
0.267 |
0.785 |
| 100 |
246249_at |
AT4G36680
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.266 |
0.129 |
| 101 |
267562_at |
AT2G39670
|
[Radical SAM superfamily protein] |
0.265 |
-0.134 |
| 102 |
265991_at |
AT2G24120
|
PDE319, PIGMENT DEFECTIVE 319, SCA3, SCABRA 3 |
0.265 |
-0.268 |
| 103 |
254147_at |
AT4G24270
|
EMB140, EMBRYO DEFECTIVE 140 |
0.264 |
-0.101 |
| 104 |
264262_at |
AT1G09200
|
H3.1, histone 3.1 |
0.264 |
0.022 |
| 105 |
260994_at |
AT1G12130
|
[Flavin-binding monooxygenase family protein] |
0.263 |
-0.056 |
| 106 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
0.260 |
0.262 |
| 107 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
0.260 |
0.184 |
| 108 |
251166_at |
AT3G63350
|
AT-HSFA7B, HSFA7B, HEAT SHOCK TRANSCRIPTION FACTOR A7B |
0.257 |
0.063 |
| 109 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
0.257 |
1.092 |
| 110 |
256764_at |
AT3G29310
|
[calmodulin-binding protein-related] |
0.257 |
-0.015 |
| 111 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.252 |
0.939 |
| 112 |
263847_at |
AT2G36970
|
[UDP-Glycosyltransferase superfamily protein] |
0.252 |
-0.253 |
| 113 |
254652_at |
AT4G18170
|
ATWRKY28, WRKY28, WRKY DNA-binding protein 28 |
0.251 |
1.135 |
| 114 |
266957_at |
AT2G34640
|
HMR, HEMERA, PTAC12, plastid transcriptionally active 12, TAC12 |
0.251 |
-0.114 |
| 115 |
253175_at |
AT4G35050
|
MSI3, MULTICOPY SUPPRESSOR OF IRA1 3, NFC3, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP C 3 |
0.251 |
-0.078 |
| 116 |
256833_at |
AT3G22910
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.251 |
0.235 |
| 117 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
0.250 |
1.153 |
| 118 |
247274_at |
AT5G64360
|
EIP9, EMF1-Interacting Protein 1 |
0.249 |
0.008 |
| 119 |
257352_at |
AT2G34900
|
GTE01, GTE1, GLOBAL TRANSCRIPTION FACTOR GROUP E1, IMB1, IMBIBITION-INDUCIBLE 1 |
0.249 |
-0.010 |
| 120 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
0.247 |
0.610 |
| 121 |
250443_at |
AT5G10520
|
RBK1, ROP binding protein kinases 1 |
0.247 |
0.330 |
| 122 |
259441_at |
AT1G02300
|
[Cysteine proteinases superfamily protein] |
0.247 |
0.091 |
| 123 |
263548_at |
AT2G21660
|
ATGRP7, GLYCINE RICH PROTEIN 7, CCR2, cold, circadian rhythm, and rna binding 2, GR-RBP7, GLYCINE-RICH RNA-BINDING PROTEIN 7, GRP7, GLYCINE-RICH RNA-BINDING PROTEIN 7 |
0.246 |
0.085 |
| 124 |
250434_at |
AT5G10390
|
H3.1, histone 3.1 |
0.245 |
0.061 |
| 125 |
261621_at |
AT1G01960
|
EDA10, embryo sac development arrest 10 |
0.245 |
-0.089 |
| 126 |
261625_at |
AT1G01930
|
[zinc finger protein-related] |
0.245 |
-0.033 |
| 127 |
264171_at |
AT1G02100
|
SBI1, SUPPRESSOR OF BRI1 |
0.245 |
0.003 |
| 128 |
248163_at |
AT5G54510
|
DFL1, DWARF IN LIGHT 1, GH3.6, Gretchen Hagen3.6 |
0.243 |
0.635 |
| 129 |
250529_at |
AT5G08610
|
PDE340, PIGMENT DEFECTIVE 340 |
0.242 |
-0.130 |
| 130 |
263741_at |
AT2G20620
|
unknown |
0.239 |
-0.087 |
| 131 |
249916_at |
AT5G22880
|
H2B, HISTONE H2B, HTB2, histone B2 |
0.239 |
-0.024 |
| 132 |
264172_at |
AT1G02120
|
VAD1, VASCULAR ASSOCIATED DEATH1 |
0.239 |
-0.006 |
| 133 |
245347_at |
AT4G14890
|
FdC1, ferredoxin C 1 |
0.238 |
-0.196 |
| 134 |
246944_at |
AT5G25450
|
[Cytochrome bd ubiquinol oxidase, 14kDa subunit] |
0.238 |
0.060 |
| 135 |
253432_at |
AT4G32450
|
MEF8S, MEF8 similar |
0.238 |
-0.152 |
| 136 |
245151_at |
AT2G47550
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.236 |
0.543 |
| 137 |
264987_at |
AT1G27030
|
unknown |
0.235 |
-0.070 |
| 138 |
264019_at |
AT2G21130
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
0.235 |
0.054 |
| 139 |
246929_at |
AT5G25210
|
unknown |
0.235 |
0.213 |
| 140 |
265796_at |
AT2G35730
|
[Heavy metal transport/detoxification superfamily protein ] |
0.234 |
0.573 |
| 141 |
254011_at |
AT4G26370
|
[antitermination NusB domain-containing protein] |
0.233 |
0.111 |
| 142 |
254630_at |
AT4G18360
|
GOX3, glycolate oxidase 3 |
0.233 |
0.553 |
| 143 |
259696_at |
AT1G63150
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.232 |
-0.120 |
| 144 |
266335_at |
AT2G32440
|
ATKAO2, ARABIDOPSIS ENT-KAURENOIC ACID HYDROXYLASE 2, CYP88A4, KAO2, ent-kaurenoic acid hydroxylase 2 |
0.232 |
-0.134 |
| 145 |
251200_at |
AT3G63010
|
ATGID1B, GID1B, GA INSENSITIVE DWARF1B |
0.232 |
0.416 |
| 146 |
258254_at |
AT3G26782
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.230 |
-0.028 |
| 147 |
249115_at |
AT5G43810
|
AGO10, ARGONAUTE 10, PNH, PINHEAD, ZLL, ZWILLE |
0.230 |
-0.336 |
| 148 |
262307_at |
AT1G71000
|
[Chaperone DnaJ-domain superfamily protein] |
0.230 |
0.015 |
| 149 |
264150_at |
AT1G02090
|
ATCSN7, ARABIDOPSIS THALIANA COP9 SIGNALOSOME SUBUNIT 7, COP15, CONSTITUTIVE PHOTOMORPHOGENIC 15, CSN7, COP9 SIGNALOSOME SUBUNIT 7, FUS5, FUSCA 5 |
0.229 |
-0.005 |
| 150 |
254014_at |
AT4G26120
|
[Ankyrin repeat family protein / BTB/POZ domain-containing protein] |
0.229 |
0.098 |
| 151 |
248605_at |
AT5G49410
|
unknown |
0.228 |
-0.046 |
| 152 |
251766_at |
AT3G55910
|
unknown |
0.228 |
0.304 |
| 153 |
253054_at |
AT4G37580
|
COP3, CONSTITUTIVE PHOTOMORPHOGENIC 3, HLS1, HOOKLESS 1, UNS2, UNUSUAL SUGAR RESPONSE 2 |
0.228 |
-0.403 |
| 154 |
259429_at |
AT1G01600
|
CYP86A4, cytochrome P450, family 86, subfamily A, polypeptide 4 |
0.228 |
0.022 |
| 155 |
254955_at |
AT4G10920
|
KELP |
0.227 |
-0.005 |
| 156 |
261534_at |
AT1G01820
|
PEX11C, peroxin 11c |
0.227 |
0.150 |
| 157 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
0.227 |
0.971 |
| 158 |
250555_at |
AT5G07950
|
unknown |
0.226 |
0.036 |
| 159 |
261559_at |
AT1G01780
|
PLIM2b, PLIM2b |
0.224 |
-0.025 |
| 160 |
262643_at |
AT1G62770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.224 |
0.072 |
| 161 |
261561_at |
AT1G01730
|
unknown |
0.224 |
0.170 |
| 162 |
264177_at |
AT1G02150
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.224 |
-0.110 |
| 163 |
248238_at |
AT5G53900
|
[Serine/threonine-protein kinase WNK (With No Lysine)-related] |
0.223 |
-0.293 |
| 164 |
264168_at |
AT1G02080
|
[transcription regulators] |
0.223 |
-0.060 |
| 165 |
248563_at |
AT5G49690
|
[UDP-Glycosyltransferase superfamily protein] |
0.222 |
0.038 |
| 166 |
264471_at |
AT1G67120
|
AtMDN1, MDN1, MIDASIN 1 |
0.222 |
-0.014 |
| 167 |
256675_at |
AT3G52170
|
[DNA binding] |
0.221 |
-0.188 |
| 168 |
260438_at |
AT1G68290
|
ENDO2, endonuclease 2 |
0.221 |
0.488 |
| 169 |
263411_at |
AT2G28710
|
[C2H2-type zinc finger family protein] |
0.221 |
0.373 |
| 170 |
253733_at |
AT4G29170
|
ATMND1 |
0.221 |
-0.025 |
| 171 |
261537_at |
AT1G01800
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.220 |
-0.027 |
| 172 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.220 |
0.244 |
| 173 |
259913_at |
AT1G72660
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.220 |
-0.105 |
| 174 |
246052_at |
AT5G08305
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.220 |
-0.013 |
| 175 |
256518_at |
AT1G66080
|
unknown |
0.220 |
-0.351 |
| 176 |
252281_at |
AT3G49320
|
[Metal-dependent protein hydrolase] |
0.219 |
0.044 |
| 177 |
265345_at |
AT2G22680
|
WAVH1, WAV3 homolog 1 |
0.218 |
0.109 |
| 178 |
267102_at |
AT2G41500
|
EMB2776, LIS, LACHESIS |
0.217 |
-0.037 |
| 179 |
250680_at |
AT5G06570
|
[alpha/beta-Hydrolases superfamily protein] |
0.217 |
0.283 |
| 180 |
257772_at |
AT3G23080
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.217 |
-0.070 |
| 181 |
260398_at |
AT1G72320
|
APUM23, pumilio 23, PUM23, pumilio 23 |
0.216 |
-0.090 |
| 182 |
266778_at |
AT2G29090
|
CYP707A2, cytochrome P450, family 707, subfamily A, polypeptide 2 |
0.216 |
-0.181 |
| 183 |
266340_at |
AT2G01480
|
[O-fucosyltransferase family protein] |
0.216 |
-0.052 |
| 184 |
260462_at |
AT1G10970
|
ATZIP4, ZIP4, zinc transporter 4 precursor |
0.216 |
-0.495 |
| 185 |
250463_at |
AT5G10030
|
OBF4, OCS ELEMENT BINDING FACTOR 4, TGA4, TGACG motif-binding factor 4 |
0.214 |
-0.105 |
| 186 |
260065_at |
AT1G73760
|
[RING/U-box superfamily protein] |
0.214 |
0.315 |
| 187 |
264712_at |
AT1G09810
|
ECT11, evolutionarily conserved C-terminal region 11 |
0.214 |
-0.231 |
| 188 |
261558_at |
AT1G01770
|
unknown |
0.214 |
-0.012 |
| 189 |
264005_at |
AT2G22470
|
AGP2, arabinogalactan protein 2, ATAGP2 |
0.213 |
0.773 |
| 190 |
250987_at |
AT5G02860
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.213 |
0.058 |
| 191 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.213 |
-0.015 |
| 192 |
251760_at |
AT3G55605
|
[Mitochondrial glycoprotein family protein] |
0.213 |
-0.075 |
| 193 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
0.212 |
-0.063 |
| 194 |
253299_at |
AT4G33800
|
unknown |
0.212 |
-0.036 |
| 195 |
256574_at |
AT3G14780
|
unknown |
0.211 |
0.024 |
| 196 |
260876_at |
AT1G21460
|
AtSWEET1, SWEET1 |
0.210 |
-0.185 |
| 197 |
249849_at |
AT5G23230
|
NIC2, nicotinamidase 2 |
0.209 |
0.170 |
| 198 |
247679_at |
AT5G59540
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.209 |
0.067 |
| 199 |
261301_at |
AT1G48570
|
[zinc finger (Ran-binding) family protein] |
0.208 |
-0.190 |
| 200 |
246418_at |
AT5G16960
|
[Zinc-binding dehydrogenase family protein] |
0.208 |
0.294 |
| 201 |
262014_at |
AT1G35660
|
unknown |
0.207 |
-0.257 |
| 202 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
0.207 |
1.304 |
| 203 |
247151_at |
AT5G65640
|
bHLH093, beta HLH protein 93 |
0.207 |
0.104 |
| 204 |
261654_at |
AT1G01920
|
[SET domain-containing protein] |
0.206 |
-0.029 |
| 205 |
254177_at |
AT4G23860
|
[PHD finger protein-related] |
0.205 |
0.008 |
| 206 |
256792_at |
AT3G22150
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.205 |
-0.496 |
| 207 |
264774_at |
AT1G22890
|
unknown |
0.204 |
0.444 |
| 208 |
256274_at |
AT3G12080
|
emb2738, embryo defective 2738 |
0.204 |
-0.133 |
| 209 |
251029_at |
AT5G02050
|
[Mitochondrial glycoprotein family protein] |
0.203 |
-0.021 |
| 210 |
263209_at |
AT1G10522
|
PRIN2, PLASTID REDOX INSENSITIVE 2 |
0.202 |
-0.069 |
| 211 |
260781_at |
AT1G14620
|
DECOY, DECOY |
0.202 |
-0.034 |
| 212 |
250724_at |
AT5G06330
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.202 |
-0.067 |
| 213 |
262549_at |
AT1G31290
|
AGO3, ARGONAUTE 3 |
0.201 |
0.022 |
| 214 |
262754_at |
AT1G16350
|
[Aldolase-type TIM barrel family protein] |
0.201 |
-0.087 |
| 215 |
260367_at |
AT1G69760
|
unknown |
0.201 |
0.032 |
| 216 |
264259_at |
AT1G09290
|
unknown |
0.200 |
-0.039 |
| 217 |
251856_at |
AT3G54720
|
AMP1, ALTERED MERISTEM PROGRAM 1, AtAMP1, COP2, CONSTITUTIVE MORPHOGENESIS 2, HPT, HAUPTLING, MFO1, Multifolia, PT, PRIMORDIA TIMING |
0.200 |
-0.348 |
| 218 |
257670_at |
AT3G20340
|
unknown |
0.199 |
0.311 |
| 219 |
261762_at |
AT1G15510
|
ATECB2, ARABIDOPSIS EARLY CHLOROPLAST BIOGENESIS2, ECB2, EARLY CHLOROPLAST BIOGENESIS2, VAC1, VANILLA CREAM 1 |
0.199 |
-0.367 |
| 220 |
252210_at |
AT3G50410
|
OBP1, OBF binding protein 1 |
0.199 |
0.320 |
| 221 |
260359_at |
AT1G69210
|
[Uncharacterised protein family UPF0090] |
0.199 |
-0.043 |
| 222 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
0.199 |
-0.157 |
| 223 |
264121_at |
AT1G02280
|
ATTOC33, PPI1, PLASTID PROTEIN IMPORT 1, TOC33, translocon at the outer envelope membrane of chloroplasts 33 |
0.198 |
-0.144 |
| 224 |
249767_at |
AT5G24090
|
ATCHIA, chitinase A, CHIA, chitinase A |
0.198 |
0.442 |
| 225 |
264689_at |
AT1G09900
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.198 |
-0.186 |
| 226 |
267148_at |
AT2G23470
|
RUS4, ROOT UV-B SENSITIVE 4 |
0.198 |
-0.253 |
| 227 |
266803_at |
AT2G28930
|
APK1B, protein kinase 1B, PK1B, protein kinase 1B |
0.197 |
-0.033 |
| 228 |
247551_at |
AT5G60980
|
[Nuclear transport factor 2 (NTF2) family protein with RNA binding (RRM-RBD-RNP motifs) domain] |
0.197 |
-0.016 |
| 229 |
259293_at |
AT3G11580
|
[AP2/B3-like transcriptional factor family protein] |
0.197 |
0.326 |
| 230 |
264571_at |
AT1G05330
|
unknown |
0.197 |
0.028 |
| 231 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
0.196 |
0.701 |
| 232 |
260608_at |
AT2G43870
|
[Pectin lyase-like superfamily protein] |
0.196 |
0.003 |
| 233 |
250649_at |
AT5G06690
|
WCRKC1, WCRKC thioredoxin 1 |
0.195 |
0.185 |
| 234 |
247995_at |
AT5G56160
|
[Sec14p-like phosphatidylinositol transfer family protein] |
0.193 |
0.329 |
| 235 |
254778_at |
AT4G12750
|
[Homeodomain-like transcriptional regulator] |
0.193 |
-0.022 |
| 236 |
246842_at |
AT5G26731
|
unknown |
0.193 |
0.204 |
| 237 |
257074_at |
AT3G19660
|
unknown |
0.193 |
0.569 |
| 238 |
247688_at |
AT5G59760
|
unknown |
0.192 |
0.670 |
| 239 |
250433_at |
AT5G10400
|
H3.1, histone 3.1 |
0.192 |
-0.003 |
| 240 |
251346_at |
AT3G60980
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.192 |
0.235 |
| 241 |
262605_at |
AT1G15170
|
[MATE efflux family protein] |
0.191 |
0.066 |
| 242 |
248299_at |
AT5G53080
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.191 |
-0.209 |
| 243 |
254663_at |
AT4G18290
|
KAT2, potassium channel in Arabidopsis thaliana 2 |
0.191 |
-0.132 |
| 244 |
259899_at |
AT1G71210
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.191 |
0.035 |
| 245 |
246238_at |
AT4G36670
|
AtPLT6, AtPMT6, PLT6, polyol transporter 6, PMT6, polyol/monosaccharide transporter 6 |
0.190 |
0.826 |
| 246 |
248547_at |
AT5G50280
|
EMB1006, embryo defective 1006 |
0.190 |
-0.253 |
| 247 |
250909_at |
AT5G03700
|
[D-mannose binding lectin protein with Apple-like carbohydrate-binding domain] |
0.189 |
0.322 |
| 248 |
252362_at |
AT3G48500
|
PDE312, PIGMENT DEFECTIVE 312, PTAC10, PLASTID TRANSCRIPTIONALLY ACTIVE 10, TAC10 |
0.189 |
-0.127 |
| 249 |
261642_at |
AT1G27680
|
APL2, ADPGLC-PPase large subunit |
0.188 |
-0.051 |
| 250 |
266076_at |
AT2G40700
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.188 |
-0.051 |
| 251 |
259918_at |
AT1G72570
|
[Integrase-type DNA-binding superfamily protein] |
-0.793 |
-0.051 |
| 252 |
263123_at |
AT1G78500
|
PEN6, PENTACYCLIC TRITERPENE SYNTHASE 6 |
-0.655 |
0.018 |
| 253 |
249548_at |
AT5G38170
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.621 |
0.008 |
| 254 |
257421_at |
AT1G12030
|
unknown |
-0.611 |
-0.375 |
| 255 |
249547_at |
AT5G38160
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.538 |
-0.018 |
| 256 |
258111_at |
AT3G14630
|
CYP72A9, cytochrome P450, family 72, subfamily A, polypeptide 9 |
-0.533 |
0.040 |
| 257 |
258580_at |
AT3G04170
|
[RmlC-like cupins superfamily protein] |
-0.532 |
0.002 |
| 258 |
254036_at |
AT4G25980
|
[Peroxidase superfamily protein] |
-0.487 |
-0.012 |
| 259 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
-0.476 |
0.065 |
| 260 |
258582_at |
AT3G04150
|
[RmlC-like cupins superfamily protein] |
-0.472 |
-0.025 |
| 261 |
246810_at |
AT5G27160
|
unknown |
-0.469 |
0.047 |
| 262 |
263942_at |
AT2G35860
|
FLA16, FASCICLIN-like arabinogalactan protein 16 precursor |
-0.460 |
-0.360 |
| 263 |
266762_at |
AT2G47120
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.460 |
0.022 |
| 264 |
265117_at |
AT1G62500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.454 |
0.212 |
| 265 |
264782_at |
AT1G08810
|
AtMYB60, myb domain protein 60, MYB60, myb domain protein 60 |
-0.448 |
-0.428 |
| 266 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
-0.444 |
-0.402 |
| 267 |
252412_at |
AT3G47295
|
unknown |
-0.444 |
-0.144 |
| 268 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
-0.443 |
0.073 |
| 269 |
263168_at |
AT1G03020
|
[Thioredoxin superfamily protein] |
-0.441 |
-0.043 |
| 270 |
245634_at |
AT1G25270
|
UMAMIT24, Usually multiple acids move in and out Transporters 24 |
-0.421 |
-0.053 |
| 271 |
249913_at |
AT5G22810
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.412 |
-0.036 |
| 272 |
246502_at |
AT5G16240
|
[Plant stearoyl-acyl-carrier-protein desaturase family protein] |
-0.411 |
-0.184 |
| 273 |
258003_at |
AT3G29030
|
ATEXP5, ARABIDOPSIS THALIANA EXPANSIN 5, ATEXPA5, ARABIDOPSIS THALIANA EXPANSIN A5, ATHEXP ALPHA 1.4, EXP5, EXPANSIN 5, EXPA5, expansin A5 |
-0.406 |
-0.240 |
| 274 |
247905_at |
AT5G57400
|
unknown |
-0.402 |
-0.031 |
| 275 |
263431_at |
AT2G22170
|
PLAT2, PLAT domain protein 2 |
-0.400 |
-0.198 |
| 276 |
253575_at |
AT4G31060
|
[Integrase-type DNA-binding superfamily protein] |
-0.399 |
-0.004 |
| 277 |
261809_at |
AT1G08340
|
[Rho GTPase activating protein with PAK-box/P21-Rho-binding domain] |
-0.398 |
-0.027 |
| 278 |
266465_at |
AT2G47750
|
GH3.9, putative indole-3-acetic acid-amido synthetase GH3.9 |
-0.388 |
-0.129 |
| 279 |
257793_at |
AT3G26960
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.388 |
-0.564 |
| 280 |
259736_at |
AT1G64390
|
AtGH9C2, glycosyl hydrolase 9C2, GH9C2, glycosyl hydrolase 9C2 |
-0.382 |
-0.185 |
| 281 |
249047_at |
AT5G44410
|
[FAD-binding Berberine family protein] |
-0.381 |
-0.611 |
| 282 |
246783_at |
AT5G27360
|
SFP2 |
-0.381 |
-0.373 |
| 283 |
253241_at |
AT4G34520
|
FAE1, FATTY ACID ELONGATION1, KCS18, 3-ketoacyl-CoA synthase 18 |
-0.380 |
0.006 |
| 284 |
247461_at |
AT5G62100
|
ATBAG2, BCL-2-associated athanogene 2, BAG2, BCL-2-associated athanogene 2 |
-0.380 |
-0.597 |
| 285 |
266770_at |
AT2G03090
|
ATEXP15, ATEXPA15, expansin A15, ATHEXP ALPHA 1.3, EXP15, EXPANSIN 15, EXPA15, expansin A15 |
-0.379 |
-0.090 |
| 286 |
258590_at |
AT3G04280
|
ARR22, response regulator 22, RR22, response regulator 22 |
-0.378 |
-0.040 |
| 287 |
251339_at |
AT3G60780
|
unknown |
-0.378 |
0.002 |
| 288 |
262446_at |
AT1G49310
|
unknown |
-0.373 |
0.001 |
| 289 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
-0.363 |
0.086 |
| 290 |
267389_at |
AT2G44460
|
BGLU28, beta glucosidase 28 |
-0.356 |
0.013 |
| 291 |
245006_at |
ATCG00340
|
PSAB |
-0.355 |
-0.022 |
| 292 |
252645_at |
AT3G44460
|
AtbZIP67, basic leucine zipper transcription factor 67, DPBF2 |
-0.353 |
0.028 |
| 293 |
261203_at |
AT1G12845
|
unknown |
-0.353 |
0.148 |
| 294 |
258578_at |
AT3G04200
|
[RmlC-like cupins superfamily protein] |
-0.351 |
0.006 |
| 295 |
260742_at |
AT1G15050
|
IAA34, indole-3-acetic acid inducible 34 |
-0.351 |
0.034 |
| 296 |
259707_at |
AT1G77490
|
TAPX, thylakoidal ascorbate peroxidase |
-0.345 |
-0.206 |
| 297 |
246315_at |
AT3G56870
|
unknown |
-0.340 |
-0.045 |
| 298 |
266712_at |
AT2G46750
|
AtGulLO2, GulLO2, L -gulono-1,4-lactone ( L -GulL) oxidase 2 |
-0.340 |
0.064 |
| 299 |
260812_at |
AT1G43650
|
UMAMIT22, Usually multiple acids move in and out Transporters 22 |
-0.339 |
-0.183 |
| 300 |
249231_at |
AT5G42030
|
ABIL4, ABL interactor-like protein 4 |
-0.333 |
-0.179 |
| 301 |
245703_at |
AT5G04380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.331 |
0.002 |
| 302 |
254572_at |
AT4G19380
|
[Long-chain fatty alcohol dehydrogenase family protein] |
-0.329 |
-0.377 |
| 303 |
248406_at |
AT5G51490
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.326 |
0.049 |
| 304 |
257271_at |
AT3G28007
|
AtSWEET4, SWEET4 |
-0.325 |
0.605 |
| 305 |
263238_at |
AT2G16580
|
SAUR8, SMALL AUXIN UPREGULATED RNA 8 |
-0.318 |
-0.047 |
| 306 |
261991_at |
AT1G33700
|
[Beta-glucosidase, GBA2 type family protein] |
-0.318 |
-0.211 |
| 307 |
252168_at |
AT3G50440
|
ATMES10, ARABIDOPSIS THALIANA METHYL ESTERASE 10, MES10, methyl esterase 10 |
-0.314 |
0.048 |
| 308 |
260136_at |
AT1G66360
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.313 |
0.018 |
| 309 |
259567_at |
AT1G20500
|
[AMP-dependent synthetase and ligase family protein] |
-0.312 |
0.006 |
| 310 |
267377_at |
AT2G26250
|
FDH, FIDDLEHEAD, KCS10, 3-ketoacyl-CoA synthase 10 |
-0.311 |
-0.120 |
| 311 |
257288_at |
AT3G29670
|
PMAT2, phenolic glucoside malonyltransferase 2 |
-0.309 |
-0.138 |
| 312 |
260267_at |
AT1G68530
|
CER6, ECERIFERUM 6, CUT1, CUTICULAR 1, G2, KCS6, 3-ketoacyl-CoA synthase 6, POP1, POLLEN-PISTIL INCOMPATIBILITY 1 |
-0.306 |
-0.025 |
| 313 |
254607_at |
AT4G18920
|
unknown |
-0.299 |
0.051 |
| 314 |
252624_at |
AT3G44735
|
ATPSK3, PHYTOSULFOKINE 3 PRECURSOR, PSK1, PSK3, PHYTOSULFOKINE 3 PRECURSOR |
-0.296 |
-0.195 |
| 315 |
249791_at |
AT5G23810
|
AAP7, amino acid permease 7 |
-0.291 |
0.193 |
| 316 |
246701_at |
AT5G28020
|
ATCYSD2, CYSTEINE SYNTHASE D2, CYSD2, cysteine synthase D2 |
-0.291 |
-0.137 |
| 317 |
248028_at |
AT5G55620
|
unknown |
-0.291 |
-0.418 |
| 318 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
-0.290 |
-0.404 |
| 319 |
251382_at |
AT3G60730
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.289 |
-0.003 |
| 320 |
265042_at |
AT1G04040
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.288 |
-0.413 |
| 321 |
246498_at |
AT5G16230
|
[Plant stearoyl-acyl-carrier-protein desaturase family protein] |
-0.287 |
-0.022 |
| 322 |
267150_at |
AT2G23510
|
SDT, spermidine disinapoyl acyltransferase |
-0.287 |
-0.012 |
| 323 |
266295_at |
AT2G29550
|
TUB7, tubulin beta-7 chain |
-0.287 |
-0.297 |
| 324 |
266849_at |
AT2G25940
|
ALPHA-VPE, alpha-vacuolar processing enzyme, ALPHAVPE |
-0.286 |
-0.034 |
| 325 |
256875_at |
AT3G26330
|
CYP71B37, cytochrome P450, family 71, subfamily B, polypeptide 37 |
-0.286 |
0.381 |
| 326 |
249705_at |
AT5G35580
|
[Protein kinase superfamily protein] |
-0.281 |
0.043 |
| 327 |
251772_at |
AT3G55920
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.278 |
0.101 |
| 328 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-0.276 |
0.224 |
| 329 |
263829_at |
AT2G40435
|
unknown |
-0.276 |
0.488 |
| 330 |
255743_at |
AT1G25375
|
[Metallo-hydrolase/oxidoreductase superfamily protein] |
-0.275 |
-0.107 |
| 331 |
266996_at |
AT2G34490
|
CYP710A2, cytochrome P450, family 710, subfamily A, polypeptide 2 |
-0.275 |
-0.076 |
| 332 |
264729_at |
AT1G22990
|
HIPP22, heavy metal associated isoprenylated plant protein 22 |
-0.275 |
-0.011 |
| 333 |
250501_at |
AT5G09640
|
SCPL19, serine carboxypeptidase-like 19, SNG2, SINAPOYLGLUCOSE ACCUMULATOR 2 |
-0.274 |
0.057 |
| 334 |
259142_at |
AT3G10200
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.274 |
-0.109 |
| 335 |
255175_at |
AT4G07960
|
ATCSLC12, Cellulose-synthase-like C12, CSLC12, CELLULOSE-SYNTHASE LIKE C12, CSLC12, Cellulose-synthase-like C12 |
-0.272 |
-0.022 |
| 336 |
265116_at |
AT1G62480
|
[Vacuolar calcium-binding protein-related] |
-0.272 |
-0.091 |
| 337 |
260262_at |
AT1G68470
|
[Exostosin family protein] |
-0.271 |
-0.105 |
| 338 |
261728_at |
AT1G76160
|
sks5, SKU5 similar 5 |
-0.270 |
0.060 |
| 339 |
266314_at |
AT2G27040
|
AGO4, ARGONAUTE 4, OCP11, OVEREXPRESSOR OF CATIONIC PEROXIDASE 11 |
-0.270 |
-0.343 |
| 340 |
250344_at |
AT5G11930
|
[Thioredoxin superfamily protein] |
-0.270 |
-0.391 |
| 341 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
-0.269 |
-0.149 |
| 342 |
257775_at |
AT3G29260
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.268 |
-0.060 |
| 343 |
267317_at |
AT2G34700
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.268 |
0.010 |
| 344 |
265994_at |
AT2G24170
|
[Endomembrane protein 70 protein family] |
-0.266 |
-0.084 |
| 345 |
265863_at |
AT2G01770
|
ATVIT1, VIT1, vacuolar iron transporter 1 |
-0.260 |
0.009 |
| 346 |
250727_at |
AT5G06430
|
[Thioredoxin superfamily protein] |
-0.259 |
-0.052 |
| 347 |
255626_at |
AT4G00780
|
[TRAF-like family protein] |
-0.258 |
0.012 |
| 348 |
265377_at |
AT2G05790
|
[O-Glycosyl hydrolases family 17 protein] |
-0.257 |
-0.342 |
| 349 |
251301_at |
AT3G61880
|
CYP78A9, cytochrome p450 78a9 |
-0.256 |
-0.052 |
| 350 |
259348_at |
AT3G03770
|
[Leucine-rich repeat protein kinase family protein] |
-0.253 |
-0.652 |
| 351 |
264704_at |
AT1G70090
|
GATL9, GALACTURONOSYLTRANSFERASE-LIKE 9, LGT8, glucosyl transferase family 8 |
-0.252 |
-0.156 |
| 352 |
252740_at |
AT3G43270
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.251 |
0.509 |
| 353 |
266035_at |
AT2G05990
|
ENR1, ENOYL-ACP REDUCTASE 1, MOD1, MOSAIC DEATH 1 |
-0.251 |
-0.162 |
| 354 |
250661_at |
AT5G07030
|
[Eukaryotic aspartyl protease family protein] |
-0.250 |
-0.199 |
| 355 |
244975_at |
ATCG00710
|
PSBH, photosystem II reaction center protein H |
-0.250 |
-0.011 |
| 356 |
267308_at |
AT2G30200
|
EMB3147, EMBRYO DEFECTIVE 3147 |
-0.249 |
-0.137 |
| 357 |
252995_at |
AT4G38370
|
[Phosphoglycerate mutase family protein] |
-0.249 |
-0.082 |
| 358 |
256960_at |
AT3G13510
|
unknown |
-0.248 |
-0.555 |
| 359 |
254928_at |
AT4G11410
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.247 |
-0.154 |
| 360 |
245004_at |
ATCG00300
|
YCF9 |
-0.246 |
-0.023 |
| 361 |
249569_at |
AT5G38070
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-0.246 |
-0.076 |
| 362 |
256825_at |
AT3G22120
|
CWLP, cell wall-plasma membrane linker protein |
-0.245 |
-0.224 |
| 363 |
257541_at |
AT3G25950
|
[TRAM, LAG1 and CLN8 (TLC) lipid-sensing domain containing protein] |
-0.245 |
-0.073 |
| 364 |
249303_at |
AT5G41460
|
unknown |
-0.245 |
-0.328 |
| 365 |
265296_at |
AT2G14060
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.245 |
-0.024 |
| 366 |
251646_at |
AT3G57780
|
unknown |
-0.245 |
-0.323 |
| 367 |
256255_at |
AT3G11280
|
[Duplicated homeodomain-like superfamily protein] |
-0.245 |
-0.116 |
| 368 |
259095_at |
AT3G05020
|
ACP, ACYL CARRIER PROTEIN, ACP1, acyl carrier protein 1 |
-0.243 |
0.002 |
| 369 |
255527_at |
AT4G02360
|
unknown |
-0.243 |
0.911 |
| 370 |
246574_at |
AT1G31670
|
[Copper amine oxidase family protein] |
-0.242 |
0.013 |
| 371 |
248911_at |
AT5G45830
|
ATDOG1, DOG1, DELAY OF GERMINATION 1, GAAS5, germination ability after storage 5, GSQ5, GLUCOSE SENSING QTL 5 |
-0.241 |
0.068 |
| 372 |
266736_at |
AT2G46960
|
CYP709B1, cytochrome P450, family 709, subfamily B, polypeptide 1 |
-0.240 |
-0.061 |
| 373 |
248510_at |
AT5G50315
|
[Mutator-like transposase family, has a 5.0e-14 P-value blast match to Q9XE24 /118-277 Pfam PF03108 MuDR family transposase (MuDr-element domain)] |
-0.240 |
0.043 |
| 374 |
264891_at |
AT1G23200
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.239 |
0.380 |
| 375 |
253619_at |
AT4G30460
|
[glycine-rich protein] |
-0.238 |
0.370 |
| 376 |
261131_at |
AT1G19835
|
unknown |
-0.235 |
-0.320 |
| 377 |
254506_at |
AT4G20140
|
GSO1, GASSHO1 |
-0.235 |
-0.092 |
| 378 |
253289_at |
AT4G34320
|
unknown |
-0.235 |
-0.068 |
| 379 |
251181_at |
AT3G62820
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.234 |
-0.031 |
| 380 |
252652_at |
AT3G44720
|
ADT4, arogenate dehydratase 4 |
-0.233 |
0.272 |
| 381 |
256673_at |
AT3G52370
|
FLA15, FASCICLIN-like arabinogalactan protein 15 precursor |
-0.233 |
-0.313 |
| 382 |
264570_at |
AT1G05350
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.233 |
0.074 |
| 383 |
251540_at |
AT3G58740
|
CSY1, citrate synthase 1 |
-0.230 |
0.035 |
| 384 |
245629_at |
AT1G56580
|
SVB, SMALLER WITH VARIABLE BRANCHES |
-0.230 |
0.119 |
| 385 |
264525_at |
AT1G10060
|
ATBCAT-1, branched-chain amino acid transaminase 1, BCAT-1, branched-chain amino acid transaminase 1, BCAT1, branched-chain amino acid transferase 1 |
-0.230 |
-0.184 |
| 386 |
253498_at |
AT4G31890
|
[ARM repeat superfamily protein] |
-0.230 |
-0.303 |
| 387 |
253387_at |
AT4G33010
|
AtGLDP1, glycine decarboxylase P-protein 1, GLDP1, glycine decarboxylase P-protein 1 |
-0.229 |
-0.156 |
| 388 |
264835_at |
AT1G03550
|
AtSCAMP4, SCAMP4, Secretory carrier membrane protein 4 |
-0.229 |
-0.033 |
| 389 |
247103_at |
AT5G66610
|
DAR7, DA1-related protein 7 |
-0.227 |
-0.052 |
| 390 |
262388_at |
AT1G49320
|
unknown |
-0.225 |
-0.030 |
| 391 |
255814_at |
AT1G19900
|
[glyoxal oxidase-related protein] |
-0.225 |
-0.012 |
| 392 |
257637_at |
AT3G25810
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.225 |
-0.012 |
| 393 |
246874_at |
AT5G26120
|
ASD2, alpha-L-arabinofuranosidase 2, ATASD2, ARABIDOPSIS THALIANA ALPHA-L-ARABINOFURANOSIDASE 2 |
-0.224 |
-0.024 |
| 394 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
-0.224 |
0.421 |
| 395 |
251898_at |
AT3G54340
|
AP3, APETALA 3, ATAP3 |
-0.223 |
0.049 |
| 396 |
257869_at |
AT3G25160
|
[ER lumen protein retaining receptor family protein] |
-0.219 |
0.015 |
| 397 |
258023_at |
AT3G19450
|
ATCAD4, CAD, CAD-C, CAD4, CINNAMYL ALCOHOL DEHYDROGENASE 4 |
-0.218 |
-0.145 |
| 398 |
250861_at |
AT5G04740
|
ACR12, ACT domain repeats 12 |
-0.217 |
-0.043 |
| 399 |
250859_at |
AT5G04660
|
CYP77A4, cytochrome P450, family 77, subfamily A, polypeptide 4 |
-0.214 |
-0.160 |
| 400 |
245310_at |
AT4G13950
|
RALFL31, ralf-like 31 |
-0.213 |
0.150 |
| 401 |
245020_at |
ATCG00540
|
PETA, photosynthetic electron transfer A |
-0.213 |
-0.047 |
| 402 |
246411_at |
AT1G57770
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
-0.213 |
-0.225 |
| 403 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
-0.213 |
-0.597 |
| 404 |
255295_at |
AT4G04760
|
[Major facilitator superfamily protein] |
-0.212 |
-0.047 |
| 405 |
248619_at |
AT5G49630
|
AAP6, amino acid permease 6 |
-0.212 |
0.081 |
| 406 |
246613_at |
AT5G35360
|
CAC2, acetyl Co-enzyme a carboxylase biotin carboxylase subunit |
-0.212 |
-0.086 |
| 407 |
249985_at |
AT5G18500
|
[Protein kinase superfamily protein] |
-0.212 |
-0.164 |
| 408 |
247770_at |
AT5G58930
|
unknown |
-0.211 |
-0.252 |
| 409 |
262525_at |
AT1G17060
|
CHI2, CHIBI 2, CYP72C1, cytochrome p450 72c1, SHK1, SHRINK 1, SOB7, SUPPRESSOR OF PHYB-4 7 |
-0.211 |
0.164 |
| 410 |
264263_at |
AT1G09155
|
AtPP2-B15, phloem protein 2-B15, PP2-B15, phloem protein 2-B15 |
-0.210 |
-0.165 |
| 411 |
246565_at |
AT5G15530
|
BCCP2, biotin carboxyl carrier protein 2, CAC1-B |
-0.210 |
-0.122 |
| 412 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
-0.210 |
0.013 |
| 413 |
245713_at |
AT5G04370
|
NAMT1 |
-0.209 |
0.105 |
| 414 |
254339_at |
AT4G22100
|
BGLU3, beta glucosidase 2 |
-0.209 |
-0.021 |
| 415 |
258485_at |
AT3G02630
|
[Plant stearoyl-acyl-carrier-protein desaturase family protein] |
-0.207 |
0.029 |
| 416 |
266635_at |
AT2G35470
|
unknown |
-0.207 |
-0.161 |
| 417 |
260644_at |
AT1G53290
|
[Galactosyltransferase family protein] |
-0.207 |
-0.077 |
| 418 |
255900_at |
AT1G17830
|
unknown |
-0.205 |
0.208 |
| 419 |
257776_at |
AT3G29190
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.205 |
0.025 |
| 420 |
265416_at |
AT2G37120
|
[S1FA-like DNA-binding protein] |
-0.205 |
0.083 |
| 421 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.204 |
-0.214 |
| 422 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.204 |
-0.505 |
| 423 |
248685_at |
AT5G48500
|
unknown |
-0.202 |
-0.145 |
| 424 |
251964_at |
AT3G53370
|
[S1FA-like DNA-binding protein] |
-0.199 |
0.274 |
| 425 |
267606_at |
AT2G26640
|
KCS11, 3-ketoacyl-CoA synthase 11 |
-0.199 |
-0.265 |
| 426 |
249497_at |
AT5G39220
|
[alpha/beta-Hydrolases superfamily protein] |
-0.199 |
-0.042 |
| 427 |
257204_at |
AT3G23805
|
RALFL24, ralf-like 24 |
-0.199 |
-0.367 |
| 428 |
262238_at |
AT1G48300
|
DGAT3, diacylglycerol acyltransferase 3 |
-0.198 |
-0.149 |
| 429 |
251402_at |
AT3G60290
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.198 |
-0.087 |
| 430 |
264562_at |
AT1G55760
|
[BTB/POZ domain-containing protein] |
-0.197 |
-0.021 |
| 431 |
255558_at |
AT4G01900
|
GLB1, GLNB1 homolog, PII |
-0.196 |
-0.013 |
| 432 |
257761_at |
AT3G23090
|
WDL3 |
-0.196 |
-0.128 |
| 433 |
247037_at |
AT5G67070
|
RALFL34, ralf-like 34 |
-0.195 |
-0.015 |
| 434 |
250258_at |
AT5G13790
|
AGL15, AGAMOUS-like 15 |
-0.192 |
-0.081 |
| 435 |
246485_at |
AT5G16080
|
AtCXE17, carboxyesterase 17, CXE17, carboxyesterase 17 |
-0.191 |
0.228 |
| 436 |
262516_at |
AT1G17190
|
ATGSTU26, glutathione S-transferase tau 26, GSTU26, glutathione S-transferase tau 26 |
-0.190 |
-0.028 |
| 437 |
255862_at |
AT2G30300
|
[Major facilitator superfamily protein] |
-0.190 |
-0.055 |
| 438 |
263628_at |
AT2G04780
|
FLA7, FASCICLIN-like arabinoogalactan 7 |
-0.190 |
-0.211 |
| 439 |
265722_at |
AT2G40100
|
LHCB4.3, light harvesting complex photosystem II |
-0.190 |
-0.513 |
| 440 |
267496_at |
AT2G30550
|
DALL3, DAD1-Like Lipase 3 |
-0.189 |
0.166 |
| 441 |
266169_at |
AT2G38900
|
[Serine protease inhibitor, potato inhibitor I-type family protein] |
-0.189 |
-0.006 |
| 442 |
261059_at |
AT1G01250
|
[Integrase-type DNA-binding superfamily protein] |
-0.188 |
-0.203 |
| 443 |
244950_at |
ATMG00160
|
COX2, cytochrome oxidase 2 |
-0.187 |
0.099 |
| 444 |
257650_at |
AT3G16800
|
[Protein phosphatase 2C family protein] |
-0.186 |
0.279 |
| 445 |
260109_at |
AT1G63260
|
TET10, tetraspanin10 |
-0.186 |
-0.285 |
| 446 |
246216_at |
AT4G36380
|
ROT3, ROTUNDIFOLIA 3 |
-0.186 |
-0.062 |
| 447 |
257573_at |
AT2G33990
|
iqd9, IQ-domain 9 |
-0.185 |
-0.185 |
| 448 |
254104_at |
AT4G25040
|
[Uncharacterised protein family (UPF0497)] |
-0.185 |
0.014 |
| 449 |
265224_at |
AT2G36710
|
[Pectin lyase-like superfamily protein] |
-0.183 |
-0.005 |
| 450 |
261570_at |
AT1G01120
|
KCS1, 3-ketoacyl-CoA synthase 1 |
-0.183 |
0.030 |
| 451 |
252169_at |
AT3G50520
|
[Phosphoglycerate mutase family protein] |
-0.182 |
0.020 |
| 452 |
251827_at |
AT3G55120
|
A11, AtCHI, CFI, CHALCONE FLAVANONE ISOMERASE, CHI, chalcone isomerase, TT5, TRANSPARENT TESTA 5 |
-0.182 |
-0.213 |
| 453 |
255248_at |
AT4G05180
|
PSBQ, PHOTOSYSTEM II SUBUNIT Q, PSBQ-2, photosystem II subunit Q-2, PSII-Q |
-0.182 |
0.011 |
| 454 |
266257_at |
AT2G27820
|
ADT3, arogenate dehydratase 3, PD1, prephenate dehydratase 1 |
-0.182 |
-0.125 |
| 455 |
257229_at |
AT3G16490
|
IQD26, IQ-domain 26 |
-0.181 |
0.125 |
| 456 |
255438_at |
AT4G03070
|
AOP, AOP1, AOP1.1 |
-0.181 |
-0.517 |
| 457 |
257938_at |
AT3G19820
|
CBB1, CABBAGE 1, DIM, DIMINUTIA, DIM1, DIMINUTO 1, DWF1, DWARF 1, EVE1, ENHANCED VERY-LOW-FLUENCE RESPONSES 1 |
-0.181 |
-0.188 |
| 458 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
-0.180 |
0.150 |
| 459 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
-0.180 |
-0.048 |
| 460 |
255302_at |
AT4G04830
|
ATMSRB5, methionine sulfoxide reductase B5, MSRB5, methionine sulfoxide reductase B5 |
-0.178 |
0.085 |
| 461 |
265617_at |
AT2G25520
|
[Drug/metabolite transporter superfamily protein] |
-0.178 |
0.045 |
| 462 |
265014_at |
AT1G24430
|
[HXXXD-type acyl-transferase family protein] |
-0.178 |
-0.026 |
| 463 |
262133_at |
AT1G78000
|
SEL1, SELENATE RESISTANT 1, SULTR1;2, sulfate transporter 1;2 |
-0.177 |
-0.007 |
| 464 |
245891_at |
AT5G09220
|
AAP2, amino acid permease 2 |
-0.176 |
-0.083 |
| 465 |
249366_at |
AT5G40610
|
GPDHp, Glycerol-3-phosphate dehydrogenase plastidic |
-0.174 |
-0.101 |
| 466 |
262608_at |
AT1G14120
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.173 |
1.010 |
| 467 |
263247_at |
AT2G31440
|
[INVOLVED IN: positive regulation of catalytic activity, protein processing] |
-0.173 |
0.009 |
| 468 |
259982_at |
AT1G76410
|
ATL8 |
-0.173 |
0.077 |
| 469 |
259527_at |
AT1G12600
|
[UDP-N-acetylglucosamine (UAA) transporter family] |
-0.172 |
-0.023 |
| 470 |
263979_at |
AT2G42840
|
PDF1, protodermal factor 1 |
-0.172 |
0.041 |
| 471 |
258258_at |
AT3G26790
|
FUS3, FUSCA 3 |
-0.172 |
-0.035 |
| 472 |
250043_at |
AT5G18430
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.172 |
-0.247 |
| 473 |
250512_at |
AT5G09995
|
unknown |
-0.172 |
-0.192 |
| 474 |
258276_at |
AT3G15710
|
[Peptidase S24/S26A/S26B/S26C family protein] |
-0.170 |
0.246 |
| 475 |
245140_at |
AT2G45420
|
LBD18, LOB domain-containing protein 18 |
-0.170 |
-0.001 |
| 476 |
260441_at |
AT1G68260
|
ALT3, acyl-lipid thioesterase 3 |
-0.170 |
-0.051 |
| 477 |
252129_at |
AT3G50890
|
AtHB28, homeobox protein 28, HB28, homeobox protein 28, ZHD7, ZINC FINGER HOMEODOMAIN 7 |
-0.169 |
-0.106 |
| 478 |
266223_at |
AT2G28790
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.169 |
0.342 |
| 479 |
256964_at |
AT3G13520
|
AGP12, arabinogalactan protein 12, ATAGP12 |
-0.168 |
-0.195 |
| 480 |
246778_at |
AT5G27450
|
MK, mevalonate kinase, MVK, MEVALONATE KINASE |
-0.167 |
-0.204 |
| 481 |
245965_at |
AT5G19730
|
[Pectin lyase-like superfamily protein] |
-0.167 |
-0.349 |
| 482 |
254145_at |
AT4G24700
|
unknown |
-0.167 |
-0.645 |
| 483 |
265066_at |
AT1G03870
|
FLA9, FASCICLIN-like arabinoogalactan 9 |
-0.167 |
-0.466 |
| 484 |
266532_at |
AT2G16890
|
[UDP-Glycosyltransferase superfamily protein] |
-0.166 |
-0.058 |
| 485 |
259904_at |
AT1G74150
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.165 |
0.007 |
| 486 |
245479_at |
AT4G16140
|
[proline-rich family protein] |
-0.164 |
-0.205 |
| 487 |
263754_at |
AT2G21510
|
[DNAJ heat shock N-terminal domain-containing protein] |
-0.164 |
0.556 |
| 488 |
247127_at |
AT5G66100
|
AtLARP1b, LARP1b, La related protein 1b |
-0.163 |
-0.003 |
| 489 |
262539_at |
AT1G17200
|
[Uncharacterised protein family (UPF0497)] |
-0.163 |
-0.260 |
| 490 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
-0.162 |
-0.650 |
| 491 |
251319_at |
AT3G61610
|
[Galactose mutarotase-like superfamily protein] |
-0.162 |
0.079 |
| 492 |
259154_at |
AT3G10260
|
[Reticulon family protein] |
-0.162 |
0.295 |
| 493 |
250041_at |
AT5G18410
|
ATSRA1, KLK, KLUNKER, PIR, PIROGI, PIR121, PIROGI 121, PIRP, SRA1 |
-0.161 |
-0.140 |
| 494 |
265038_at |
AT1G03920
|
[Protein kinase family protein] |
-0.161 |
-0.047 |
| 495 |
253984_at |
AT4G26590
|
ATOPT5, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 5, OPT5, oligopeptide transporter 5 |
-0.161 |
-0.116 |
| 496 |
267376_at |
AT2G26330
|
ER, ERECTA, QRP1, QUANTITATIVE RESISTANCE TO PLECTOSPHAERELLA 1 |
-0.161 |
-0.293 |
| 497 |
248592_at |
AT5G49280
|
[hydroxyproline-rich glycoprotein family protein] |
-0.161 |
0.440 |
| 498 |
265471_at |
AT2G37130
|
[Peroxidase superfamily protein] |
-0.161 |
0.376 |
| 499 |
244929_at |
ATMG00580
|
NAD4, NADH dehydrogenase subunit 4 |
-0.161 |
0.017 |