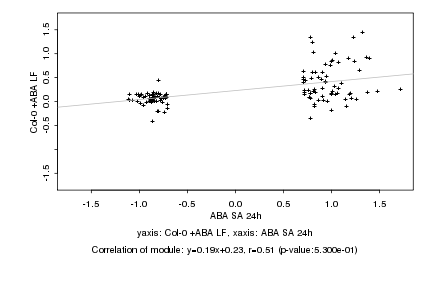
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ABA SA 24h |
Log2 signal ratio
Col-0 +ABA LF |
| 1 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
1.714 |
0.258 |
| 2 |
261221_at |
AT1G19960
|
unknown |
1.475 |
0.212 |
| 3 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
1.386 |
0.909 |
| 4 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
1.371 |
0.195 |
| 5 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.355 |
0.939 |
| 6 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.322 |
1.455 |
| 7 |
265672_at |
AT2G31980
|
AtCYS2, PHYTOCYSTATIN 2, CYS2, PHYTOCYSTATIN 2 |
1.290 |
0.653 |
| 8 |
264861_at |
AT1G24320
|
[Six-hairpin glycosidases superfamily protein] |
1.254 |
0.049 |
| 9 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
1.233 |
0.840 |
| 10 |
254413_at |
AT4G21440
|
ATM4, A. THALIANA MYB 4, ATMYB102, MYB-like 102, MYB102, MYB102, MYB-like 102 |
1.223 |
1.342 |
| 11 |
258064_at |
AT3G14680
|
CYP72A14, cytochrome P450, family 72, subfamily A, polypeptide 14 |
1.203 |
0.074 |
| 12 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
1.193 |
0.182 |
| 13 |
266415_at |
AT2G38530
|
cdf3, cell growth defect factor-3, LP2, LTP2, lipid transfer protein 2 |
1.178 |
0.155 |
| 14 |
262448_at |
AT1G49450
|
[Transducin/WD40 repeat-like superfamily protein] |
1.169 |
0.916 |
| 15 |
250366_at |
AT5G11420
|
unknown |
1.146 |
-0.091 |
| 16 |
259009_at |
AT3G09260
|
BGLU23, LEB, LONG ER BODY, PSR3.1, PYK10 |
1.141 |
0.044 |
| 17 |
260904_at |
AT1G02450
|
NIMIN-1, NIMIN1, NIM1-interacting 1 |
1.103 |
0.385 |
| 18 |
257673_at |
AT3G20370
|
[TRAF-like family protein] |
1.067 |
0.287 |
| 19 |
247727_at |
AT5G59490
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
1.063 |
0.823 |
| 20 |
261323_at |
AT1G44760
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
1.060 |
0.176 |
| 21 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
1.037 |
0.148 |
| 22 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
1.032 |
1.017 |
| 23 |
255032_at |
AT4G09500
|
[UDP-Glycosyltransferase superfamily protein] |
1.024 |
0.328 |
| 24 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
1.008 |
0.212 |
| 25 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
1.006 |
0.874 |
| 26 |
257480_at |
AT1G15640
|
unknown |
0.999 |
0.162 |
| 27 |
247902_at |
AT5G57350
|
AHA3, H(+)-ATPase 3, ATAHA3, ARABIDOPSIS THALIANA ARABIDOPSIS H(+)-ATPASE, HA3, H(+)-ATPase 3 |
0.997 |
-0.174 |
| 28 |
247421_at |
AT5G62800
|
[Protein with RING/U-box and TRAF-like domains] |
0.996 |
0.175 |
| 29 |
267559_at |
AT2G45570
|
CYP76C2, cytochrome P450, family 76, subfamily C, polypeptide 2 |
0.990 |
0.837 |
| 30 |
260039_at |
AT1G68795
|
CLE12, CLAVATA3/ESR-RELATED 12 |
0.983 |
0.768 |
| 31 |
257810_at |
AT3G27070
|
TOM20-1, translocase outer membrane 20-1 |
0.956 |
0.018 |
| 32 |
265122_at |
AT1G62540
|
FMO GS-OX2, flavin-monooxygenase glucosinolate S-oxygenase 2 |
0.944 |
0.528 |
| 33 |
246215_at |
AT4G37180
|
[Homeodomain-like superfamily protein] |
0.935 |
0.435 |
| 34 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.935 |
0.404 |
| 35 |
255723_at |
AT3G29575
|
AFP3, ABI five binding protein 3 |
0.931 |
0.789 |
| 36 |
249754_at |
AT5G24530
|
DMR6, DOWNY MILDEW RESISTANT 6 |
0.913 |
0.033 |
| 37 |
247657_at |
AT5G59845
|
[Gibberellin-regulated family protein] |
0.902 |
0.618 |
| 38 |
267496_at |
AT2G30550
|
DALL3, DAD1-Like Lipase 3 |
0.900 |
0.285 |
| 39 |
254697_at |
AT4G17970
|
ALMT12, aluminum-activated, malate transporter 12, ATALMT12, QUAC1, quick-activating anion channel 1 |
0.899 |
0.107 |
| 40 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
0.886 |
0.475 |
| 41 |
250690_at |
AT5G06530
|
ABCG22, ATP-binding cassette G22, AtABCG22, Arabidopsis thaliana ATP-binding cassette G22 |
0.860 |
0.039 |
| 42 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
0.857 |
0.510 |
| 43 |
258037_at |
AT3G21230
|
4CL5, 4-coumarate:CoA ligase 5 |
0.827 |
0.207 |
| 44 |
252073_at |
AT3G51750
|
unknown |
0.827 |
0.617 |
| 45 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
0.821 |
-0.104 |
| 46 |
260567_at |
AT2G43820
|
ATSAGT1, Arabidopsis thaliana salicylic acid glucosyltransferase 1, GT, SAGT1, salicylic acid glucosyltransferase 1, SGT1, UDP-glucose:salicylic acid glucosyltransferase 1, UGT74F2, UDP-glucosyltransferase 74F2 |
0.818 |
-0.042 |
| 47 |
245993_at |
AT5G20700
|
unknown |
0.814 |
0.264 |
| 48 |
247038_at |
AT5G67160
|
EPS1, ENHANCED PSEUDOMONAS SUSCEPTIBILTY 1 |
0.811 |
0.217 |
| 49 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
0.811 |
1.043 |
| 50 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
0.801 |
1.250 |
| 51 |
259040_at |
AT3G09270
|
ATGSTU8, glutathione S-transferase TAU 8, GSTU8, glutathione S-transferase TAU 8 |
0.798 |
0.622 |
| 52 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
0.786 |
0.483 |
| 53 |
257798_at |
AT3G15950
|
NAI2 |
0.779 |
-0.350 |
| 54 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
0.778 |
1.358 |
| 55 |
249757_at |
AT5G24316
|
[proline-rich family protein] |
0.774 |
0.074 |
| 56 |
245885_at |
AT5G09440
|
EXL4, EXORDIUM like 4 |
0.774 |
0.186 |
| 57 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
0.773 |
0.170 |
| 58 |
260278_at |
AT1G80590
|
ATWRKY66, WRKY66, WRKY DNA-binding protein 66 |
0.763 |
0.099 |
| 59 |
245277_at |
AT4G15550
|
IAGLU, indole-3-acetate beta-D-glucosyltransferase |
0.762 |
0.102 |
| 60 |
249941_at |
AT5G22270
|
unknown |
0.758 |
0.232 |
| 61 |
253017_at |
AT4G37970
|
ATCAD6, CAD6, cinnamyl alcohol dehydrogenase 6 |
0.728 |
0.451 |
| 62 |
264532_at |
AT1G55740
|
AtSIP1, seed imbibition 1, RS1, raffinose synthase 1, SIP1, seed imbibition 1 |
0.713 |
0.151 |
| 63 |
255521_at |
AT4G02280
|
ATSUS3, SUS3, sucrose synthase 3 |
0.713 |
0.246 |
| 64 |
249033_at |
AT5G44920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.709 |
0.207 |
| 65 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
0.707 |
0.645 |
| 66 |
251019_at |
AT5G02420
|
unknown |
0.707 |
0.471 |
| 67 |
264894_at |
AT1G23040
|
[hydroxyproline-rich glycoprotein family protein] |
0.705 |
0.515 |
| 68 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.704 |
0.401 |
| 69 |
255460_at |
AT4G02800
|
unknown |
-1.119 |
0.049 |
| 70 |
263607_at |
AT2G16270
|
unknown |
-1.111 |
0.025 |
| 71 |
256926_at |
AT3G22540
|
unknown |
-1.103 |
0.151 |
| 72 |
248963_at |
AT5G45700
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-1.073 |
0.034 |
| 73 |
264763_at |
AT1G61450
|
unknown |
-1.032 |
0.153 |
| 74 |
254233_at |
AT4G23800
|
3xHMG-box2, 3xHigh Mobility Group-box2 |
-1.024 |
0.015 |
| 75 |
252078_at |
AT3G51740
|
IMK2, inflorescence meristem receptor-like kinase 2 |
-1.010 |
0.153 |
| 76 |
252763_at |
AT3G42725
|
[Putative membrane lipoprotein] |
-1.006 |
0.106 |
| 77 |
247425_at |
AT5G62550
|
unknown |
-0.991 |
-0.039 |
| 78 |
255265_at |
AT4G05190
|
ATK5, kinesin 5 |
-0.989 |
0.126 |
| 79 |
257740_at |
AT3G27330
|
[zinc finger (C3HC4-type RING finger) family protein] |
-0.981 |
0.153 |
| 80 |
252148_at |
AT3G51280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.966 |
0.091 |
| 81 |
250228_at |
AT5G13840
|
CCS52B, cell cycle switch protein 52 B, FZR3, FIZZY-related 3 |
-0.959 |
-0.070 |
| 82 |
247962_at |
AT5G56580
|
ANQ1, ARABIDOPSIS NQK1, ATMKK6, ARABIDOPSIS THALIANA MAP KINASE KINASE 6, MKK6, MAP kinase kinase 6 |
-0.938 |
0.117 |
| 83 |
248057_at |
AT5G55520
|
unknown |
-0.934 |
-0.019 |
| 84 |
259930_at |
AT1G34355
|
ATPS1, PARALLEL SPINDLE 1, PS1, PARALLEL SPINDLE 1 |
-0.916 |
0.179 |
| 85 |
258464_at |
AT3G17360
|
POK1, phragmoplast orienting kinesin 1 |
-0.901 |
0.070 |
| 86 |
262109_at |
AT1G02730
|
ATCSLD5, cellulose synthase-like D5, CSLD5, CELLULOSE SYNTHASE LIKE D5, CSLD5, cellulose synthase-like D5, SOS6, SALT OVERLY SENSITIVE 6 |
-0.897 |
0.020 |
| 87 |
259686_at |
AT1G63100
|
[GRAS family transcription factor] |
-0.896 |
0.126 |
| 88 |
259151_at |
AT3G10310
|
[P-loop nucleoside triphosphate hydrolases superfamily protein with CH (Calponin Homology) domain] |
-0.894 |
0.056 |
| 89 |
248055_at |
AT5G55830
|
LecRK-S.7, L-type lectin receptor kinase S.7 |
-0.889 |
0.028 |
| 90 |
252691_at |
AT3G44050
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.875 |
-0.000 |
| 91 |
261840_at |
AT1G16070
|
AtTLP8, tubby like protein 8, TLP8, tubby like protein 8 |
-0.873 |
0.150 |
| 92 |
265085_at |
AT1G03780
|
AtTPX2, TPX2, targeting protein for XKLP2 |
-0.869 |
0.136 |
| 93 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
-0.868 |
-0.397 |
| 94 |
259851_at |
AT1G72250
|
[Di-glucose binding protein with Kinesin motor domain] |
-0.868 |
0.007 |
| 95 |
267618_at |
AT2G26760
|
CYCB1;4, Cyclin B1;4 |
-0.868 |
0.153 |
| 96 |
261780_at |
AT1G76310
|
CYCB2;4, CYCLIN B2;4 |
-0.867 |
0.050 |
| 97 |
253978_at |
AT4G26660
|
unknown |
-0.867 |
0.053 |
| 98 |
259290_at |
AT3G11520
|
CYC2, CYCLIN 2, CYCB1;3, CYCLIN B1;3 |
-0.866 |
0.089 |
| 99 |
262802_at |
AT1G20930
|
CDKB2;2, cyclin-dependent kinase B2;2 |
-0.863 |
0.077 |
| 100 |
247603_at |
AT5G60930
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.863 |
0.031 |
| 101 |
267636_at |
AT2G42110
|
unknown |
-0.860 |
0.163 |
| 102 |
264377_at |
AT2G25060
|
AtENODL14, ENODL14, early nodulin-like protein 14 |
-0.856 |
0.095 |
| 103 |
247039_at |
AT5G67270
|
ATEB1C, MICROTUBULE END BINDING PROTEIN 1, ATEB1H1, ATEB1-HOMOLOG1, EB1C, end binding protein 1C |
-0.856 |
0.199 |
| 104 |
253403_at |
AT4G32830
|
AtAUR1, ataurora1, AUR1, ataurora1 |
-0.853 |
0.165 |
| 105 |
246415_at |
AT5G17160
|
unknown |
-0.845 |
0.007 |
| 106 |
253148_at |
AT4G35620
|
CYCB2;2, Cyclin B2;2 |
-0.843 |
0.093 |
| 107 |
257115_at |
AT3G20150
|
[Kinesin motor family protein] |
-0.837 |
0.095 |
| 108 |
254964_at |
AT4G11080
|
3xHMG-box1, 3xHigh Mobility Group-box1 |
-0.829 |
0.015 |
| 109 |
251067_at |
AT5G01910
|
unknown |
-0.826 |
0.178 |
| 110 |
262081_at |
AT1G59540
|
ZCF125 |
-0.823 |
0.015 |
| 111 |
245688_at |
AT1G28290
|
AGP31, arabinogalactan protein 31 |
-0.820 |
-0.196 |
| 112 |
245739_at |
AT1G44110
|
CYCA1;1, Cyclin A1;1 |
-0.812 |
0.111 |
| 113 |
252089_at |
AT3G52110
|
unknown |
-0.811 |
0.446 |
| 114 |
248619_at |
AT5G49630
|
AAP6, amino acid permease 6 |
-0.805 |
-0.197 |
| 115 |
258067_at |
AT3G25980
|
AtMAD2, MAD2, MITOTIC ARREST-DEFICIENT 2 |
-0.804 |
0.188 |
| 116 |
264815_at |
AT1G03620
|
[ELMO/CED-12 family protein] |
-0.794 |
0.104 |
| 117 |
256864_at |
AT3G23890
|
ATTOPII, TOPII, topoisomerase II |
-0.788 |
0.037 |
| 118 |
253051_at |
AT4G37490
|
CYC1, CYCLIN 1, CYCB1, CYCB1;1, CYCLIN B1;1 |
-0.782 |
0.165 |
| 119 |
258376_at |
AT3G17680
|
[Kinase interacting (KIP1-like) family protein] |
-0.777 |
0.077 |
| 120 |
261660_at |
AT1G18370
|
ATNACK1, ARABIDOPSIS NPK1-ACTIVATING KINESIN 1, HIK, HINKEL, NACK1, NPK1-ACTIVATING KINESIN 1 |
-0.762 |
-0.014 |
| 121 |
260032_at |
AT1G68750
|
ATPPC4, phosphoenolpyruvate carboxylase 4, PPC4, phosphoenolpyruvate carboxylase 4 |
-0.759 |
0.073 |
| 122 |
248413_at |
AT5G51600
|
ATMAP65-3, ARABIDOPSIS THALIANA MICROTUBULE-ASSOCIATED PROTEIN 65-3, MAP65-3, MICROTUBULE-ASSOCIATED PROTEIN 65-3, PLE, PLEIADE |
-0.759 |
0.092 |
| 123 |
259978_at |
AT1G76540
|
CDKB2;1, cyclin-dependent kinase B2;1 |
-0.750 |
0.082 |
| 124 |
248691_at |
AT5G48310
|
unknown |
-0.744 |
0.116 |
| 125 |
261198_at |
AT1G12940
|
ATNRT2.5, nitrate transporter2.5, NRT2.5, nitrate transporter2.5 |
-0.744 |
-0.222 |
| 126 |
245607_at |
AT4G14330
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.736 |
0.145 |
| 127 |
249644_at |
AT5G37010
|
unknown |
-0.735 |
0.113 |
| 128 |
250386_at |
AT5G11510
|
AtMYB3R4, myb domain protein 3R4, MYB3R-4, myb domain protein 3r-4 |
-0.733 |
0.124 |
| 129 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
-0.731 |
0.076 |
| 130 |
261364_at |
AT1G53140
|
DRP5A, Dynamin related protein 5A |
-0.725 |
0.164 |
| 131 |
258796_at |
AT3G04630
|
WDL1, WVD2-like 1 |
-0.725 |
0.150 |
| 132 |
255410_at |
AT4G03100
|
[Rho GTPase activating protein with PAK-box/P21-Rho-binding domain] |
-0.724 |
0.091 |
| 133 |
253757_at |
AT4G28950
|
ARAC7, Arabidopsis RAC-like 7, ATRAC7, ATROP9, RAC7, ROP9, RHO-related protein from plants 9 |
-0.719 |
0.148 |
| 134 |
258471_at |
AT3G06030
|
ANP3, NPK1-related protein kinase 3, AtANP3, MAPKKK12, NP3, NPK1-related protein kinase 3 |
-0.716 |
-0.127 |
| 135 |
253804_at |
AT4G28230
|
unknown |
-0.714 |
-0.044 |