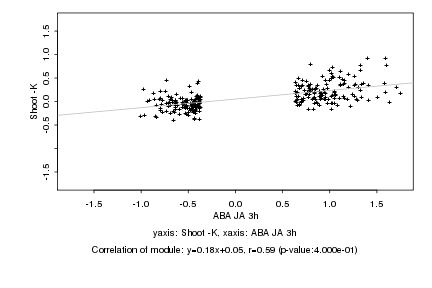
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ABA JA 3h |
Log2 signal ratio
Shoot -K |
| 1 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
1.746 |
0.178 |
| 2 |
265119_at |
AT1G62570
|
FMO GS-OX4, flavin-monooxygenase glucosinolate S-oxygenase 4 |
1.704 |
0.311 |
| 3 |
254413_at |
AT4G21440
|
ATM4, A. THALIANA MYB 4, ATMYB102, MYB-like 102, MYB102, MYB102, MYB-like 102 |
1.625 |
-0.013 |
| 4 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.598 |
0.786 |
| 5 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
1.584 |
0.925 |
| 6 |
262448_at |
AT1G49450
|
[Transducin/WD40 repeat-like superfamily protein] |
1.581 |
0.194 |
| 7 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
1.570 |
0.391 |
| 8 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
1.506 |
0.094 |
| 9 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
1.407 |
0.347 |
| 10 |
249771_at |
AT5G24080
|
[Protein kinase superfamily protein] |
1.402 |
0.036 |
| 11 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
1.397 |
0.921 |
| 12 |
265024_at |
AT1G24600
|
unknown |
1.355 |
0.405 |
| 13 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
1.336 |
0.366 |
| 14 |
251824_at |
AT3G55090
|
ABCG16, ATP-binding cassette G16 |
1.329 |
0.106 |
| 15 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
1.325 |
0.670 |
| 16 |
260357_at |
AT1G69260
|
AFP1, ABI five binding protein |
1.321 |
0.240 |
| 17 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
1.318 |
0.784 |
| 18 |
258957_at |
AT3G01420
|
ALPHA-DOX1, alpha-dioxygenase 1, DIOX1, DOX1, PADOX-1, plant alpha dioxygenase 1 |
1.300 |
0.302 |
| 19 |
261243_at |
AT1G20180
|
unknown |
1.285 |
0.035 |
| 20 |
256464_at |
AT1G32560
|
AtLEA4-1, Late Embryogenesis Abundant 4-1, LEA4-1, Late Embryogenesis Abundant 4-1 |
1.274 |
0.049 |
| 21 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
1.268 |
0.373 |
| 22 |
266462_at |
AT2G47770
|
ATTSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related, TSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related |
1.261 |
0.535 |
| 23 |
251580_at |
AT3G58450
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
1.257 |
0.107 |
| 24 |
263073_at |
AT2G17500
|
[Auxin efflux carrier family protein] |
1.257 |
0.358 |
| 25 |
257540_at |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
1.234 |
0.154 |
| 26 |
266503_at |
AT2G47780
|
[Rubber elongation factor protein (REF)] |
1.219 |
-0.103 |
| 27 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
1.195 |
0.303 |
| 28 |
258322_at |
AT3G22740
|
HMT3, homocysteine S-methyltransferase 3 |
1.193 |
0.580 |
| 29 |
263825_at |
AT2G40370
|
LAC5, laccase 5 |
1.183 |
0.047 |
| 30 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
1.151 |
0.067 |
| 31 |
256789_at |
AT3G13672
|
SINA2 |
1.148 |
0.396 |
| 32 |
253101_at |
AT4G37430
|
CYP81F1, CYTOCHROME P450 MONOOXYGENASE 81F1, CYP91A2, cytochrome P450, family 91, subfamily A, polypeptide 2 |
1.147 |
0.465 |
| 33 |
265796_at |
AT2G35730
|
[Heavy metal transport/detoxification superfamily protein ] |
1.141 |
0.363 |
| 34 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
1.139 |
0.118 |
| 35 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.128 |
0.484 |
| 36 |
259705_at |
AT1G77450
|
anac032, NAC domain containing protein 32, NAC032, NAC domain containing protein 32 |
1.111 |
0.656 |
| 37 |
264436_at |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
1.106 |
0.346 |
| 38 |
265913_at |
AT2G25625
|
unknown |
1.105 |
0.377 |
| 39 |
245982_at |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
1.102 |
0.527 |
| 40 |
248227_at |
AT5G53820
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.098 |
0.069 |
| 41 |
256069_at |
AT1G13740
|
AFP2, ABI five binding protein 2 |
1.079 |
-0.079 |
| 42 |
264102_at |
AT1G79270
|
ECT8, evolutionarily conserved C-terminal region 8 |
1.055 |
0.168 |
| 43 |
256757_at |
AT3G25620
|
ABCG21, ATP-binding cassette G21 |
1.054 |
0.143 |
| 44 |
265216_at |
AT1G05100
|
MAPKKK18, mitogen-activated protein kinase kinase kinase 18 |
1.050 |
0.229 |
| 45 |
265188_at |
AT1G23800
|
ALDH2B, aldehyde dehydrogenase 2B, ALDH2B7, aldehyde dehydrogenase 2B7 |
1.047 |
0.078 |
| 46 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
1.044 |
0.198 |
| 47 |
258732_at |
AT3G05820
|
A/N-InvH, alkaline/neutral invertase H, At-A/N-InvH, Arabidopsis alkaline/neutral invertase H, INVH, invertase H |
1.041 |
-0.021 |
| 48 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
1.034 |
0.525 |
| 49 |
267026_at |
AT2G38340
|
DREB19, dehydration response element-binding protein 19 |
1.029 |
0.548 |
| 50 |
245748_at |
AT1G51140
|
AKS1, ABA-responsive kinase substrate 1, FBH3, FLOWERING BHLH 3 |
1.028 |
-0.027 |
| 51 |
267147_at |
AT2G38240
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
1.024 |
0.132 |
| 52 |
260040_at |
AT1G68765
|
IDA, INFLORESCENCE DEFICIENT IN ABSCISSION |
1.024 |
0.726 |
| 53 |
262616_at |
AT1G06620
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
1.018 |
0.509 |
| 54 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
1.017 |
0.600 |
| 55 |
249941_at |
AT5G22270
|
unknown |
1.012 |
-0.022 |
| 56 |
261570_at |
AT1G01120
|
KCS1, 3-ketoacyl-CoA synthase 1 |
1.012 |
-0.154 |
| 57 |
245160_at |
AT2G33080
|
AtRLP28, receptor like protein 28, RLP28, receptor like protein 28 |
1.011 |
0.112 |
| 58 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
0.995 |
0.468 |
| 59 |
247487_at |
AT5G62150
|
[peptidoglycan-binding LysM domain-containing protein] |
0.994 |
0.673 |
| 60 |
266143_at |
AT2G38905
|
[Low temperature and salt responsive protein family] |
0.986 |
0.045 |
| 61 |
259384_at |
AT3G16450
|
JAL33, Jacalin-related lectin 33 |
0.975 |
-0.022 |
| 62 |
260101_at |
AT1G73260
|
ATKTI1, ARABIDOPSIS THALIANA KUNITZ TRYPSIN INHIBITOR 1, KTI1, kunitz trypsin inhibitor 1 |
0.972 |
0.289 |
| 63 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.963 |
0.366 |
| 64 |
247421_at |
AT5G62800
|
[Protein with RING/U-box and TRAF-like domains] |
0.954 |
0.090 |
| 65 |
250292_at |
AT5G13220
|
JAS1, JASMONATE-ASSOCIATED 1, JAZ10, jasmonate-zim-domain protein 10, TIFY9, TIFY DOMAIN PROTEIN 9 |
0.954 |
0.448 |
| 66 |
255250_at |
AT4G05100
|
AtMYB74, myb domain protein 74, MYB74, myb domain protein 74 |
0.953 |
0.186 |
| 67 |
256378_at |
AT1G66830
|
[Leucine-rich repeat protein kinase family protein] |
0.946 |
0.058 |
| 68 |
255521_at |
AT4G02280
|
ATSUS3, SUS3, sucrose synthase 3 |
0.934 |
0.199 |
| 69 |
245386_at |
AT4G14010
|
RALFL32, ralf-like 32 |
0.934 |
0.060 |
| 70 |
251668_at |
AT3G57010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.927 |
0.267 |
| 71 |
250252_at |
AT5G13750
|
ZIFL1, zinc induced facilitator-like 1 |
0.919 |
0.135 |
| 72 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.914 |
0.549 |
| 73 |
265359_at |
AT2G16720
|
ATMYB7, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 7, ATY49, MYB7, myb domain protein 7 |
0.905 |
0.175 |
| 74 |
260841_at |
AT1G29195
|
unknown |
0.902 |
0.078 |
| 75 |
249063_at |
AT5G44110
|
ABCI21, ATP-binding cassette A21, ATNAP2, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN, ATPOP1, POP1 |
0.901 |
0.058 |
| 76 |
247957_at |
AT5G57050
|
ABI2, ABA INSENSITIVE 2, AtABI2 |
0.895 |
0.183 |
| 77 |
250882_at |
AT5G04000
|
unknown |
0.893 |
0.120 |
| 78 |
248352_at |
AT5G52300
|
LTI65, LOW-TEMPERATURE-INDUCED 65, RD29B, RESPONSIVE TO DESSICATION 29B |
0.886 |
0.231 |
| 79 |
245724_at |
AT1G73390
|
[Endosomal targeting BRO1-like domain-containing protein] |
0.885 |
-0.076 |
| 80 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
0.885 |
0.231 |
| 81 |
265618_at |
AT2G25460
|
unknown |
0.884 |
0.109 |
| 82 |
263482_at |
AT2G03980
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.870 |
-0.039 |
| 83 |
253203_at |
AT4G34710
|
ADC2, arginine decarboxylase 2, ATADC2, SPE2 |
0.868 |
0.350 |
| 84 |
267496_at |
AT2G30550
|
DALL3, DAD1-Like Lipase 3 |
0.859 |
0.280 |
| 85 |
259380_at |
AT3G16340
|
ABCG29, ATP-binding cassette G29, AtABCG29, ATPDR1, PLEIOTROPIC DRUG RESISTANCE 1, PDR1, pleiotropic drug resistance 1 |
0.858 |
-0.009 |
| 86 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
0.848 |
0.286 |
| 87 |
258880_at |
AT3G06420
|
ATG8H, autophagy 8h |
0.845 |
0.266 |
| 88 |
261220_at |
AT1G19970
|
[ER lumen protein retaining receptor family protein] |
0.844 |
0.055 |
| 89 |
253606_at |
AT4G30530
|
GGP1, gamma-glutamyl peptidase 1 |
0.838 |
0.095 |
| 90 |
259439_at |
AT1G01480
|
ACS2, 1-amino-cyclopropane-1-carboxylate synthase 2, AT-ACC2 |
0.837 |
0.206 |
| 91 |
254269_at |
AT4G23050
|
[PAS domain-containing protein tyrosine kinase family protein] |
0.836 |
-0.037 |
| 92 |
265084_at |
AT1G03790
|
AtTZF4, SOM, SOMNUS, TZF4, Tandem CCCH Zinc Finger protein 4 |
0.835 |
0.173 |
| 93 |
245264_at |
AT4G17245
|
[RING/U-box superfamily protein] |
0.827 |
-0.153 |
| 94 |
262282_at |
AT1G68610
|
PCR11, PLANT CADMIUM RESISTANCE 11 |
0.822 |
0.067 |
| 95 |
253619_at |
AT4G30460
|
[glycine-rich protein] |
0.815 |
0.274 |
| 96 |
256725_at |
AT2G34070
|
TBL37, TRICHOME BIREFRINGENCE-LIKE 37 |
0.809 |
0.063 |
| 97 |
260264_at |
AT1G68500
|
unknown |
0.787 |
0.259 |
| 98 |
256324_at |
AT1G66760
|
[MATE efflux family protein] |
0.787 |
0.223 |
| 99 |
247727_at |
AT5G59490
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.787 |
0.372 |
| 100 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
0.786 |
0.797 |
| 101 |
267032_at |
AT2G38490
|
CIPK22, CBL-interacting protein kinase 22, SnRK3.19, SNF1-RELATED PROTEIN KINASE 3.19 |
0.784 |
0.154 |
| 102 |
260144_at |
AT1G71960
|
ABCG25, ATP-binding casette G25, ATABCG25, Arabidopsis thaliana ATP-binding cassette G25 |
0.783 |
0.243 |
| 103 |
248700_at |
AT5G48400
|
ATGLR1.2, GLR1.2, GLUTAMATE RECEPTOR 1.2 |
0.775 |
0.286 |
| 104 |
267080_at |
AT2G41190
|
[Transmembrane amino acid transporter family protein] |
0.774 |
0.346 |
| 105 |
251191_at |
AT3G62590
|
[alpha/beta-Hydrolases superfamily protein] |
0.771 |
0.336 |
| 106 |
262933_at |
AT1G65840
|
ATPAO4, polyamine oxidase 4, PAO4, polyamine oxidase 4 |
0.768 |
-0.162 |
| 107 |
254390_at |
AT4G21940
|
CPK15, calcium-dependent protein kinase 15 |
0.765 |
0.098 |
| 108 |
255462_at |
AT4G02940
|
[oxidoreductase, 2OG-Fe(II) oxygenase family protein] |
0.762 |
0.200 |
| 109 |
245209_at |
AT5G12340
|
unknown |
0.759 |
0.203 |
| 110 |
251644_at |
AT3G57540
|
[Remorin family protein] |
0.753 |
0.439 |
| 111 |
264252_at |
AT1G09180
|
ATSAR1, SECRETION-ASSOCIATED RAS 1, ATSARA1A, secretion-associated RAS super family 1, SARA1A, secretion-associated RAS super family 1 |
0.744 |
0.157 |
| 112 |
265122_at |
AT1G62540
|
FMO GS-OX2, flavin-monooxygenase glucosinolate S-oxygenase 2 |
0.736 |
0.167 |
| 113 |
249134_at |
AT5G43150
|
unknown |
0.727 |
0.252 |
| 114 |
265573_at |
AT2G28200
|
[C2H2-type zinc finger family protein] |
0.714 |
0.031 |
| 115 |
253219_at |
AT4G34990
|
AtMYB32, myb domain protein 32, MYB32, myb domain protein 32 |
0.714 |
0.084 |
| 116 |
247435_at |
AT5G62480
|
ATGSTU9, glutathione S-transferase tau 9, GST14, GLUTATHIONE S-TRANSFERASE 14, GST14B, GLUTATHIONE S-TRANSFERASE 14B, GSTU9, glutathione S-transferase tau 9 |
0.711 |
0.356 |
| 117 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.704 |
0.466 |
| 118 |
250483_at |
AT5G10300
|
AtHNL, ATMES5, ARABIDOPSIS THALIANA METHYL ESTERASE 5, HNL, HYDROXYNITRILE LYASE, MES5, methyl esterase 5 |
0.694 |
0.333 |
| 119 |
258293_at |
AT3G23430
|
ATPHO1, ARABIDOPSIS PHOSPHATE 1, PHO1, phosphate 1 |
0.693 |
-0.011 |
| 120 |
256073_at |
AT1G18100
|
E12A11, MFT, MOTHER OF FT AND TFL1 |
0.693 |
0.046 |
| 121 |
250246_at |
AT5G13700
|
APAO, ATPAO1, polyamine oxidase 1, PAO1, polyamine oxidase 1 |
0.690 |
0.007 |
| 122 |
248701_at |
AT5G48410
|
ATGLR1.3, ARABIDOPSIS THALIANA GLUTAMATE RECEPTOR 1.3, GLR1.3, glutamate receptor 1.3 |
0.678 |
0.289 |
| 123 |
262751_at |
AT1G16310
|
[Cation efflux family protein] |
0.678 |
-0.052 |
| 124 |
257954_at |
AT3G21760
|
HYR1, HYPOSTATIN RESISTANCE 1 |
0.676 |
-0.045 |
| 125 |
259616_at |
AT1G47960
|
ATC/VIF1, CELL WALL / VACUOLAR INHIBITOR OF FRUCTOSIDASE 1, C/VIF1, cell wall / vacuolar inhibitor of fructosidase 1 |
0.675 |
-0.064 |
| 126 |
266486_at |
AT2G47950
|
unknown |
0.671 |
0.284 |
| 127 |
254189_at |
AT4G24000
|
ATCSLG2, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G2, CSLG2, cellulose synthase like G2 |
0.665 |
0.507 |
| 128 |
263318_at |
AT2G24762
|
AtGDU4, glutamine dumper 4, GDU4, glutamine dumper 4 |
0.662 |
0.121 |
| 129 |
259297_at |
AT3G05360
|
AtRLP30, receptor like protein 30, RLP30, receptor like protein 30 |
0.659 |
0.347 |
| 130 |
250501_at |
AT5G09640
|
SCPL19, serine carboxypeptidase-like 19, SNG2, SINAPOYLGLUCOSE ACCUMULATOR 2 |
0.656 |
0.056 |
| 131 |
263709_at |
AT1G09310
|
unknown |
0.652 |
-0.077 |
| 132 |
247175_at |
AT5G65280
|
GCL1, GCR2-like 1 |
0.649 |
0.202 |
| 133 |
253458_at |
AT4G32070
|
Phox4, Phox4 |
0.646 |
-0.008 |
| 134 |
259445_at |
AT1G02400
|
ATGA2OX4, Arabidopsis thaliana gibberellin 2-oxidase 4, ATGA2OX6, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 6, DTA1, DOWNSTREAM TARGET OF AGL15 1, GA2OX6, gibberellin 2-oxidase 6 |
0.644 |
0.356 |
| 135 |
250149_at |
AT5G14700
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.640 |
0.121 |
| 136 |
254304_at |
AT4G22270
|
ATMRB1, ARABIDOPSIS MEMBRANE RELATED BIGGER1, MRB1, MEMBRANE RELATED BIGGER1 |
0.639 |
0.030 |
| 137 |
256149_at |
AT1G55110
|
AtIDD7, indeterminate(ID)-domain 7, IDD7, indeterminate(ID)-domain 7 |
0.636 |
0.009 |
| 138 |
254188_at |
AT4G23920
|
ATUGE2, UDP-GLC 4-EPIMERASE 2, UGE2, UDP-D-glucose/UDP-D-galactose 4-epimerase 2 |
0.634 |
0.188 |
| 139 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
0.634 |
0.408 |
| 140 |
256883_at |
AT3G26440
|
unknown |
0.633 |
0.229 |
| 141 |
251899_at |
AT3G54400
|
[Eukaryotic aspartyl protease family protein] |
-1.011 |
-0.304 |
| 142 |
267384_at |
AT2G44370
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.978 |
0.266 |
| 143 |
257891_at |
AT3G17170
|
RFC3, REGULATOR OF FATTY-ACID COMPOSITION 3 |
-0.970 |
-0.279 |
| 144 |
246366_at |
AT1G51850
|
[Leucine-rich repeat protein kinase family protein] |
-0.937 |
0.011 |
| 145 |
267034_at |
AT2G38310
|
PYL4, PYR1-like 4, RCAR10, regulatory components of ABA receptor 10 |
-0.917 |
0.040 |
| 146 |
262388_at |
AT1G49320
|
unknown |
-0.872 |
0.181 |
| 147 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
-0.867 |
0.053 |
| 148 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
-0.858 |
-0.317 |
| 149 |
261662_at |
AT1G18350
|
ATMKK7, MAP kinase kinase 7, BUD1, BUSHY AND DWARF 1, MKK7, MAP kinase kinase 7, MKK7, MAP KINASE KINASE7 |
-0.845 |
-0.065 |
| 150 |
246001_at |
AT5G20790
|
unknown |
-0.840 |
-0.331 |
| 151 |
251491_at |
AT3G59480
|
[pfkB-like carbohydrate kinase family protein] |
-0.812 |
0.045 |
| 152 |
253218_at |
AT4G34980
|
SLP2, subtilisin-like serine protease 2 |
-0.805 |
-0.144 |
| 153 |
267385_at |
AT2G44380
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.804 |
0.229 |
| 154 |
249151_at |
AT5G43360
|
ATPT4, PHT1;3, phosphate transporter 1;3, PHT3, PHOSPHATE TRANSPORTER 3 |
-0.799 |
0.082 |
| 155 |
250444_at |
AT5G10560
|
[Glycosyl hydrolase family protein] |
-0.797 |
-0.175 |
| 156 |
262001_at |
AT1G33790
|
[jacalin lectin family protein] |
-0.787 |
-0.051 |
| 157 |
263599_at |
AT2G01830
|
AHK4, ARABIDOPSIS HISTIDINE KINASE 4, ATCRE1, CRE1, CYTOKININ RESPONSE 1, WOL, WOODEN LEG, WOL1, WOODEN LEG 1 |
-0.777 |
-0.230 |
| 158 |
252176_at |
AT3G50720
|
[Protein kinase superfamily protein] |
-0.777 |
0.040 |
| 159 |
258270_at |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
-0.743 |
0.225 |
| 160 |
261762_at |
AT1G15510
|
ATECB2, ARABIDOPSIS EARLY CHLOROPLAST BIOGENESIS2, ECB2, EARLY CHLOROPLAST BIOGENESIS2, VAC1, VANILLA CREAM 1 |
-0.736 |
-0.201 |
| 161 |
254280_at |
AT4G22756
|
ATSMO1-2, SMO1-2, sterol C4-methyl oxidase 1-2 |
-0.734 |
-0.030 |
| 162 |
252234_at |
AT3G49780
|
ATPSK3 (FORMER SYMBOL), ATPSK4, phytosulfokine 4 precursor, PSK4, phytosulfokine 4 precursor |
-0.733 |
0.450 |
| 163 |
256890_at |
AT3G23830
|
AtGRP4, GR-RBP4, GRP4, glycine-rich RNA-binding protein 4 |
-0.717 |
0.113 |
| 164 |
253777_at |
AT4G28450
|
[nucleotide binding] |
-0.717 |
-0.036 |
| 165 |
251771_at |
AT3G56000
|
ATCSLA14, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE A14, CSLA14, cellulose synthase like A14 |
-0.715 |
-0.094 |
| 166 |
251900_at |
AT3G54430
|
SRS6, SHI-related sequence 6 |
-0.708 |
-0.005 |
| 167 |
263194_at |
AT1G36060
|
[Integrase-type DNA-binding superfamily protein] |
-0.700 |
-0.002 |
| 168 |
255852_at |
AT1G66970
|
GDPDL1, Glycerophosphodiester phosphodiesterase (GDPD) like 1, SVL2, SHV3-like 2 |
-0.697 |
-0.240 |
| 169 |
259021_at |
AT3G07540
|
[Actin-binding FH2 (formin homology 2) family protein] |
-0.697 |
-0.081 |
| 170 |
246077_at |
AT5G20420
|
CHR42, chromatin remodeling 42 |
-0.693 |
0.060 |
| 171 |
263276_at |
AT2G14100
|
CYP705A13, cytochrome P450, family 705, subfamily A, polypeptide 13 |
-0.682 |
0.097 |
| 172 |
250860_at |
AT5G04770
|
ATCAT6, ARABIDOPSIS THALIANA CATIONIC AMINO ACID TRANSPORTER 6, CAT6, cationic amino acid transporter 6 |
-0.673 |
-0.008 |
| 173 |
250300_at |
AT5G11890
|
EMB3135, EMBRYO DEFECTIVE 3135 |
-0.668 |
0.024 |
| 174 |
250529_at |
AT5G08610
|
PDE340, PIGMENT DEFECTIVE 340 |
-0.666 |
-0.199 |
| 175 |
266957_at |
AT2G34640
|
HMR, HEMERA, PTAC12, plastid transcriptionally active 12, TAC12 |
-0.662 |
-0.400 |
| 176 |
253650_at |
AT4G30020
|
[PA-domain containing subtilase family protein] |
-0.655 |
-0.221 |
| 177 |
253087_at |
AT4G36350
|
ATPAP25, ARABIDOPSIS THALIANA PURPLE ACID PHOSPHATASE 25, PAP25, purple acid phosphatase 25 |
-0.646 |
0.043 |
| 178 |
250694_at |
AT5G06710
|
HAT14, homeobox from Arabidopsis thaliana |
-0.646 |
-0.070 |
| 179 |
266546_at |
AT2G35270
|
AHL21, AT-hook motif nuclear localized protein 21, GIK, GIANT KILLER |
-0.645 |
0.026 |
| 180 |
245676_at |
AT1G56670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.640 |
0.006 |
| 181 |
259672_at |
AT1G68990
|
MGP3, male gametophyte defective 3 |
-0.640 |
-0.032 |
| 182 |
251310_at |
AT3G61150
|
HD-GL2-1, HOMEODOMAIN-GLABRA2 1, HDG1, homeodomain GLABROUS 1 |
-0.638 |
-0.109 |
| 183 |
263679_at |
AT1G59990
|
EMB3108, EMBRYO DEFECTIVE 3108, HS3, HEAVY SEED 3, RH22, RNA helicase 22 |
-0.638 |
-0.154 |
| 184 |
252716_at |
AT3G43920
|
ATDCL3, DICER-LIKE 3, DCL3, dicer-like 3 |
-0.633 |
-0.020 |
| 185 |
247297_at |
AT5G64100
|
[Peroxidase superfamily protein] |
-0.626 |
0.161 |
| 186 |
251287_at |
AT3G61820
|
[Eukaryotic aspartyl protease family protein] |
-0.619 |
-0.084 |
| 187 |
262849_at |
AT1G14710
|
[hydroxyproline-rich glycoprotein family protein] |
-0.598 |
-0.169 |
| 188 |
256168_at |
AT1G51805
|
[Leucine-rich repeat protein kinase family protein] |
-0.596 |
-0.269 |
| 189 |
250059_at |
AT5G17820
|
[Peroxidase superfamily protein] |
-0.586 |
0.083 |
| 190 |
245644_at |
AT1G25320
|
[Leucine-rich repeat protein kinase family protein] |
-0.567 |
-0.125 |
| 191 |
247946_at |
AT5G57180
|
CIA2, chloroplast import apparatus 2 |
-0.566 |
0.068 |
| 192 |
249097_at |
AT5G43520
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.563 |
-0.019 |
| 193 |
258856_at |
AT3G02040
|
AtGDPD1, GDPD1, Glycerophosphodiester phosphodiesterase 1, SRG3, senescence-related gene 3 |
-0.562 |
-0.054 |
| 194 |
252624_at |
AT3G44735
|
ATPSK3, PHYTOSULFOKINE 3 PRECURSOR, PSK1, PSK3, PHYTOSULFOKINE 3 PRECURSOR |
-0.559 |
-0.035 |
| 195 |
251667_at |
AT3G57150
|
AtCBF5, AtNAP57, CBF5, NAP57, homologue of NAP57 |
-0.553 |
-0.050 |
| 196 |
265864_at |
AT2G01750
|
ATMAP70-3, microtubule-associated proteins 70-3, MAP70-3, microtubule-associated proteins 70-3 |
-0.542 |
-0.127 |
| 197 |
266314_at |
AT2G27040
|
AGO4, ARGONAUTE 4, OCP11, OVEREXPRESSOR OF CATIONIC PEROXIDASE 11 |
-0.537 |
-0.090 |
| 198 |
266976_at |
AT2G39410
|
[alpha/beta-Hydrolases superfamily protein] |
-0.533 |
0.004 |
| 199 |
263438_at |
AT2G28660
|
[Chloroplast-targeted copper chaperone protein] |
-0.533 |
0.022 |
| 200 |
255888_at |
AT1G20300
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.532 |
-0.135 |
| 201 |
253566_at |
AT4G31210
|
[DNA topoisomerase, type IA, core] |
-0.531 |
-0.238 |
| 202 |
247444_at |
AT5G62630
|
HIPL2, hipl2 protein precursor |
-0.528 |
-0.081 |
| 203 |
246184_at |
AT5G20950
|
[Glycosyl hydrolase family protein] |
-0.527 |
-0.017 |
| 204 |
258051_at |
AT3G16260
|
TRZ4, tRNAse Z4 |
-0.526 |
-0.275 |
| 205 |
259444_at |
AT1G02370
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.526 |
-0.085 |
| 206 |
259211_at |
AT3G09020
|
[alpha 1,4-glycosyltransferase family protein] |
-0.523 |
0.030 |
| 207 |
246478_at |
AT5G15980
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.523 |
-0.079 |
| 208 |
256062_at |
AT1G07090
|
LSH6, LIGHT SENSITIVE HYPOCOTYLS 6 |
-0.519 |
0.033 |
| 209 |
266891_at |
AT2G44690
|
ARAC9, Arabidopsis RAC-like 9, ATROP8, RHO-RELATED PROTEIN FROM PLANTS 8, ROP8, RHO-RELATED PROTEIN FROM PLANTS 8 |
-0.518 |
0.028 |
| 210 |
256796_at |
AT3G22210
|
unknown |
-0.514 |
-0.134 |
| 211 |
254631_at |
AT4G18610
|
LSH9, LIGHT SENSITIVE HYPOCOTYLS 9 |
-0.511 |
0.043 |
| 212 |
259822_at |
AT1G66230
|
AtMYB20, myb domain protein 20, MYB20, myb domain protein 20 |
-0.505 |
-0.224 |
| 213 |
262648_at |
AT1G14030
|
LSMT-L, lysine methyltransferase (LSMT)-like |
-0.504 |
-0.286 |
| 214 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
-0.492 |
0.327 |
| 215 |
253156_at |
AT4G35730
|
[Regulator of Vps4 activity in the MVB pathway protein] |
-0.488 |
-0.139 |
| 216 |
248507_at |
AT5G50420
|
[O-fucosyltransferase family protein] |
-0.487 |
-0.179 |
| 217 |
248472_at |
AT5G50860
|
[Protein kinase superfamily protein] |
-0.484 |
-0.026 |
| 218 |
248092_at |
AT5G55170
|
ATSUMO3, SUM3, SMALL UBIQUITIN-LIKE MODIFIER 3, SUMO 3, SMALL UBIQUITIN-LIKE MODIFIER 3, SUMO3, small ubiquitin-like modifier 3 |
-0.474 |
0.193 |
| 219 |
254079_at |
AT4G25730
|
[FtsJ-like methyltransferase family protein] |
-0.469 |
-0.163 |
| 220 |
247352_at |
AT5G63650
|
SNRK2-5, SNRK2.5, SNF1-related protein kinase 2.5, SRK2H, SNF1-RELATED PROTEIN KINASE 2H |
-0.469 |
-0.146 |
| 221 |
258385_at |
AT3G15510
|
ANAC056, Arabidopsis NAC domain containing protein 56, ATNAC2, NAC domain containing protein 2, NAC2, NAC domain containing protein 2, NARS1, NAC-REGULATED SEED MORPHOLOGY 1 |
-0.468 |
0.059 |
| 222 |
257150_at |
AT3G27230
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.467 |
-0.074 |
| 223 |
262586_at |
AT1G15480
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.464 |
-0.029 |
| 224 |
259082_at |
AT3G04820
|
[Pseudouridine synthase family protein] |
-0.463 |
-0.085 |
| 225 |
250761_at |
AT5G06050
|
[Putative methyltransferase family protein] |
-0.461 |
-0.179 |
| 226 |
262170_at |
AT1G74940
|
unknown |
-0.460 |
-0.103 |
| 227 |
257054_at |
AT3G15353
|
ATMT3, MT3, metallothionein 3 |
-0.457 |
-0.026 |
| 228 |
265961_at |
AT2G37400
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.455 |
-0.112 |
| 229 |
260685_at |
AT1G17650
|
AtGLYR2, GLYR2, glyoxylate reductase 2, GR2, GLYOXYLATE REDUCTASE 2 |
-0.453 |
-0.199 |
| 230 |
245117_at |
AT2G41560
|
ACA4, autoinhibited Ca(2+)-ATPase, isoform 4 |
-0.450 |
-0.222 |
| 231 |
261603_at |
AT1G49600
|
ATRBP47A, RNA-binding protein 47A, RBP47A, RNA-binding protein 47A |
-0.449 |
0.000 |
| 232 |
247191_at |
AT5G65310
|
ATHB-5, ATHB5, homeobox protein 5, HB5, homeobox protein 5 |
-0.449 |
-0.223 |
| 233 |
251143_at |
AT5G01220
|
SQD2, sulfoquinovosyldiacylglycerol 2 |
-0.441 |
-0.342 |
| 234 |
252305_at |
AT3G49240
|
emb1796, embryo defective 1796 |
-0.440 |
-0.060 |
| 235 |
250613_at |
AT5G07240
|
IQD24, IQ-domain 24 |
-0.437 |
-0.372 |
| 236 |
245770_at |
AT1G30240
|
[FUNCTIONS IN: binding] |
-0.437 |
0.034 |
| 237 |
261814_at |
AT1G08310
|
[alpha/beta-Hydrolases superfamily protein] |
-0.436 |
-0.081 |
| 238 |
253745_at |
AT4G29090
|
[Ribonuclease H-like superfamily protein] |
-0.436 |
0.005 |
| 239 |
252254_at |
AT3G49250
|
DMS3, DEFECTIVE IN MERISTEM SILENCING 3, IDN1, INVOLVED IN DE NOVO 1 |
-0.435 |
-0.099 |
| 240 |
255288_at |
AT4G04670
|
[Met-10+ like family protein / kelch repeat-containing protein] |
-0.432 |
-0.092 |
| 241 |
247826_at |
AT5G58480
|
[O-Glycosyl hydrolases family 17 protein] |
-0.432 |
-0.158 |
| 242 |
251083_at |
AT5G01590
|
TIC56, TRANSLOCON AT THE INNER ENVELOPE MEMBRANE OF CHLOROPLASTS 56 |
-0.431 |
-0.255 |
| 243 |
246457_at |
AT5G16750
|
TOZ, TORMOZEMBRYO DEFECTIVE |
-0.431 |
-0.057 |
| 244 |
266924_at |
AT2G45730
|
[eukaryotic initiation factor 3 gamma subunit family protein] |
-0.430 |
-0.096 |
| 245 |
259316_at |
AT3G01175
|
unknown |
-0.429 |
0.049 |
| 246 |
250419_at |
AT5G11250
|
[Disease resistance protein (TIR-NBS-LRR class)] |
-0.428 |
-0.180 |
| 247 |
252511_at |
AT3G46280
|
[protein kinase-related] |
-0.427 |
0.020 |
| 248 |
266608_at |
AT2G35500
|
SKL2, shikimate kinase-like 2 |
-0.427 |
-0.120 |
| 249 |
262888_at |
AT1G14790
|
AtRDR1, ATRDRP1, RDR1, RNA-dependent RNA polymerase 1 |
-0.425 |
-0.193 |
| 250 |
266254_at |
AT2G27810
|
ATNAT12, ARABIDOPSIS NUCLEOBASE-ASCORBATE TRANSPORTER 12, NAT12, nucleobase-ascorbate transporter 12 |
-0.421 |
-0.123 |
| 251 |
260882_at |
AT1G29280
|
ATWRKY65, WRKY DNA-BINDING PROTEIN 65, WRKY65, WRKY DNA-binding protein 65 |
-0.419 |
0.116 |
| 252 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
-0.412 |
0.384 |
| 253 |
266358_at |
AT2G32280
|
VCC, VASCULATURE COMPLEXITY AND CONNECTIVITY |
-0.408 |
0.145 |
| 254 |
255491_at |
AT4G02670
|
AtIDD12, indeterminate(ID)-domain 12, IDD12, indeterminate(ID)-domain 12 |
-0.408 |
0.069 |
| 255 |
252852_at |
AT4G39900
|
unknown |
-0.405 |
-0.071 |
| 256 |
252990_at |
AT4G38440
|
IYO, MINIYO |
-0.405 |
0.072 |
| 257 |
251519_at |
AT3G59400
|
GUN4, GENOMES UNCOUPLED 4 |
-0.404 |
-0.105 |
| 258 |
247643_at |
AT5G60450
|
ARF4, auxin response factor 4 |
-0.402 |
-0.044 |
| 259 |
249019_at |
AT5G44780
|
MORF4, multiple organellar RNA editing factor 4 |
-0.399 |
-0.001 |
| 260 |
260037_at |
AT1G68840
|
AtRAV2, EDF2, ETHYLENE RESPONSE DNA BINDING FACTOR 2, RAP2.8, RELATED TO AP2 8, RAV2, related to ABI3/VP1 2, TEM2, TEMPRANILLO 2 |
-0.397 |
0.045 |
| 261 |
260991_at |
AT1G12160
|
[Flavin-binding monooxygenase family protein] |
-0.396 |
0.047 |
| 262 |
264471_at |
AT1G67120
|
AtMDN1, MDN1, MIDASIN 1 |
-0.396 |
-0.105 |
| 263 |
266482_at |
AT2G47640
|
[Small nuclear ribonucleoprotein family protein] |
-0.392 |
0.093 |
| 264 |
248298_at |
AT5G53110
|
[RING/U-box superfamily protein] |
-0.392 |
0.434 |
| 265 |
257741_at |
AT3G27340
|
unknown |
-0.392 |
-0.015 |
| 266 |
266803_at |
AT2G28930
|
APK1B, protein kinase 1B, PK1B, protein kinase 1B |
-0.391 |
-0.146 |
| 267 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
-0.390 |
-0.092 |
| 268 |
262263_at |
AT1G70940
|
ATPIN3, ARABIDOPSIS PIN-FORMED 3, PIN3, PIN-FORMED 3 |
-0.389 |
-0.212 |
| 269 |
260292_at |
AT1G63680
|
APG13, ALBINO OR PALE-GREEN 13, ATMURE, MURE, PDE316, PIGMENT DEFECTIVE EMBRYO 316 |
-0.387 |
-0.379 |
| 270 |
245984_at |
AT5G13090
|
unknown |
-0.386 |
0.040 |
| 271 |
267462_at |
AT2G33735
|
[Chaperone DnaJ-domain superfamily protein] |
-0.385 |
0.008 |
| 272 |
254778_at |
AT4G12750
|
[Homeodomain-like transcriptional regulator] |
-0.383 |
-0.034 |
| 273 |
249948_at |
AT5G19210
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.382 |
-0.026 |
| 274 |
266076_at |
AT2G40700
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.382 |
0.052 |
| 275 |
248254_at |
AT5G53320
|
[Leucine-rich repeat protein kinase family protein] |
-0.382 |
-0.005 |
| 276 |
248405_at |
AT5G51480
|
SKS2, SKU5 similar 2 |
-0.381 |
0.125 |
| 277 |
263962_at |
AT2G36350
|
[Protein kinase superfamily protein] |
-0.377 |
-0.115 |
| 278 |
266684_at |
AT2G19720
|
rps15ab, ribosomal protein S15A B |
-0.375 |
0.096 |
| 279 |
259863_at |
AT1G72630
|
ELF4-L2, ELF4-like 2 |
-0.375 |
0.055 |