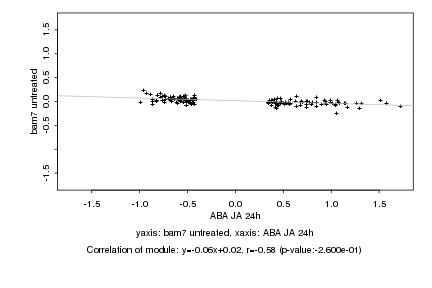
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ABA JA 24h |
Log2 signal ratio
bam7 untreated |
| 1 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
1.719 |
-0.090 |
| 2 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
1.573 |
-0.025 |
| 3 |
259384_at |
AT3G16450
|
JAL33, Jacalin-related lectin 33 |
1.514 |
0.033 |
| 4 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
1.309 |
-0.021 |
| 5 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
1.291 |
-0.143 |
| 6 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
1.254 |
-0.036 |
| 7 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
1.166 |
-0.105 |
| 8 |
245885_at |
AT5G09440
|
EXL4, EXORDIUM like 4 |
1.142 |
-0.025 |
| 9 |
262616_at |
AT1G06620
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
1.133 |
-0.032 |
| 10 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
1.085 |
-0.032 |
| 11 |
246215_at |
AT4G37180
|
[Homeodomain-like superfamily protein] |
1.069 |
-0.020 |
| 12 |
267612_at |
AT2G26690
|
AtNPF6.2, NPF6.2, NRT1/ PTR family 6.2 |
1.065 |
0.034 |
| 13 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
1.049 |
-0.232 |
| 14 |
246963_at |
AT5G24820
|
[Eukaryotic aspartyl protease family protein] |
1.042 |
-0.065 |
| 15 |
250292_at |
AT5G13220
|
JAS1, JASMONATE-ASSOCIATED 1, JAZ10, jasmonate-zim-domain protein 10, TIFY9, TIFY DOMAIN PROTEIN 9 |
1.037 |
-0.058 |
| 16 |
260101_at |
AT1G73260
|
ATKTI1, ARABIDOPSIS THALIANA KUNITZ TRYPSIN INHIBITOR 1, KTI1, kunitz trypsin inhibitor 1 |
1.007 |
-0.022 |
| 17 |
260546_at |
AT2G43520
|
ATTI2, trypsin inhibitor protein 2, TI2, trypsin inhibitor protein 2 |
0.990 |
0.028 |
| 18 |
266330_at |
AT2G01530
|
MLP329, MLP-like protein 329, ZCE2, (Zusammen-CA)-enhanced 2 |
0.983 |
-0.018 |
| 19 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.946 |
-0.060 |
| 20 |
245244_at |
AT1G44350
|
ILL6, IAA-leucine resistant (ILR)-like gene 6 |
0.939 |
-0.034 |
| 21 |
264887_at |
AT1G23120
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.932 |
0.005 |
| 22 |
247266_at |
AT5G64570
|
ATBXL4, ARABIDOPSIS THALIANA BETA-D-XYLOSIDASE 4, XYL4, beta-D-xylosidase 4 |
0.927 |
0.040 |
| 23 |
267147_at |
AT2G38240
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.888 |
-0.061 |
| 24 |
252698_at |
AT3G43670
|
[Copper amine oxidase family protein] |
0.839 |
0.085 |
| 25 |
252137_at |
AT3G50980
|
XERO1, dehydrin xero 1 |
0.838 |
-0.088 |
| 26 |
256725_at |
AT2G34070
|
TBL37, TRICHOME BIREFRINGENCE-LIKE 37 |
0.837 |
-0.007 |
| 27 |
246984_at |
AT5G67310
|
CYP81G1, cytochrome P450, family 81, subfamily G, polypeptide 1 |
0.802 |
-0.001 |
| 28 |
263482_at |
AT2G03980
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.786 |
-0.055 |
| 29 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
0.767 |
-0.007 |
| 30 |
266269_at |
AT2G29480
|
ATGSTU2, glutathione S-transferase tau 2, GST20, GLUTATHIONE S-TRANSFERASE 20, GSTU2, glutathione S-transferase tau 2 |
0.751 |
0.008 |
| 31 |
261285_at |
AT1G35720
|
ANN1, annexin 1, ANNAT1, annexin 1, AtANN1, ATOXY5, OXY5 |
0.739 |
0.017 |
| 32 |
256324_at |
AT1G66760
|
[MATE efflux family protein] |
0.737 |
-0.054 |
| 33 |
258362_at |
AT3G14280
|
unknown |
0.733 |
-0.121 |
| 34 |
251996_at |
AT3G52840
|
BGAL2, beta-galactosidase 2 |
0.699 |
-0.013 |
| 35 |
253629_at |
AT4G30450
|
[glycine-rich protein] |
0.680 |
0.005 |
| 36 |
253373_at |
AT4G33150
|
LKR, LKR/SDH, LYSINE-KETOGLUTARATE REDUCTASE/SACCHAROPINE DEHYDROGENASE, SDH, SACCHAROPINE DEHYDROGENASE |
0.675 |
-0.074 |
| 37 |
261033_at |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
0.672 |
-0.006 |
| 38 |
251743_at |
AT3G55890
|
[Yippee family putative zinc-binding protein] |
0.629 |
-0.099 |
| 39 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
0.629 |
0.124 |
| 40 |
251235_at |
AT3G62860
|
[alpha/beta-Hydrolases superfamily protein] |
0.626 |
0.018 |
| 41 |
261785_at |
AT1G08230
|
[Transmembrane amino acid transporter family protein] |
0.568 |
0.061 |
| 42 |
247193_at |
AT5G65380
|
[MATE efflux family protein] |
0.565 |
-0.030 |
| 43 |
262096_at |
AT1G56010
|
anac021, Arabidopsis NAC domain containing protein 21, ANAC022, Arabidopsis NAC domain containing protein 22, NAC1, NAC domain containing protein 1 |
0.560 |
-0.054 |
| 44 |
255016_at |
AT4G10120
|
ATSPS4F |
0.556 |
-0.057 |
| 45 |
253534_at |
AT4G31500
|
ATR4, ALTERED TRYPTOPHAN REGULATION 4, CYP83B1, cytochrome P450, family 83, subfamily B, polypeptide 1, RED1, RED ELONGATED 1, RNT1, RUNT 1, SUR2, SUPERROOT 2 |
0.550 |
-0.017 |
| 46 |
250483_at |
AT5G10300
|
AtHNL, ATMES5, ARABIDOPSIS THALIANA METHYL ESTERASE 5, HNL, HYDROXYNITRILE LYASE, MES5, methyl esterase 5 |
0.524 |
-0.026 |
| 47 |
252915_at |
AT4G38810
|
[Calcium-binding EF-hand family protein] |
0.515 |
-0.006 |
| 48 |
251427_at |
AT3G60130
|
BGLU16, beta glucosidase 16 |
0.506 |
-0.047 |
| 49 |
247356_at |
AT5G63800
|
BGAL6, beta-galactosidase 6, MUM2, MUCILAGE-MODIFIED 2 |
0.498 |
-0.027 |
| 50 |
262607_at |
AT1G13990
|
unknown |
0.478 |
0.011 |
| 51 |
260943_at |
AT1G45145
|
ATH5, THIOREDOXIN H-TYPE 5, ATTRX5, thioredoxin H-type 5, LIV1, LOCUS OF INSENSITIVITY TO VICTORIN 1, TRX-h5, THIOREDOXIN H-TYPE 5, TRX5, thioredoxin H-type 5 |
0.469 |
-0.029 |
| 52 |
256061_at |
AT1G07040
|
unknown |
0.468 |
-0.025 |
| 53 |
258751_at |
AT3G05890
|
RCI2B, RARE-COLD-INDUCIBLE 2B |
0.466 |
0.067 |
| 54 |
252993_at |
AT4G38540
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
0.462 |
-0.002 |
| 55 |
250937_at |
AT5G03230
|
unknown |
0.441 |
-0.068 |
| 56 |
254573_at |
AT4G19420
|
[Pectinacetylesterase family protein] |
0.436 |
-0.072 |
| 57 |
265345_at |
AT2G22680
|
WAVH1, WAV3 homolog 1 |
0.435 |
-0.049 |
| 58 |
260887_at |
AT1G29160
|
[Dof-type zinc finger DNA-binding family protein] |
0.430 |
0.075 |
| 59 |
260986_at |
AT1G53580
|
ETHE1, ETHE1-LIKE, GLX2-3, GLYOXALASE 2-3, GLY3, glyoxalase II 3 |
0.426 |
0.007 |
| 60 |
255463_at |
AT4G02960
|
ATRE2, retro element 2, RE2, retro element 2 |
0.419 |
-0.126 |
| 61 |
246620_at |
AT5G36220
|
CYP81D1, cytochrome P450, family 81, subfamily D, polypeptide 1, CYP91A1, CYTOCHROME P450 91A1 |
0.418 |
-0.112 |
| 62 |
258727_at |
AT3G11930
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.418 |
-0.040 |
| 63 |
259154_at |
AT3G10260
|
[Reticulon family protein] |
0.402 |
-0.013 |
| 64 |
252097_at |
AT3G51090
|
unknown |
0.398 |
0.053 |
| 65 |
247601_at |
AT5G60850
|
OBP4, OBF binding protein 4 |
0.398 |
-0.003 |
| 66 |
248263_at |
AT5G53370
|
ATPMEPCRF, PECTIN METHYLESTERASE PCR FRAGMENT F, PMEPCRF, pectin methylesterase PCR fragment F |
0.386 |
-0.010 |
| 67 |
246510_at |
AT5G15410
|
ATCNGC2, CYCLIC NUCLEOTIDE-GATED CHANNEL 2, CNGC2, CYCLIC NUCLEOTIDE GATED CHANNEL 2, DND1, DEFENSE NO DEATH 1 |
0.384 |
-0.011 |
| 68 |
266479_at |
AT2G31160
|
LSH3, LIGHT SENSITIVE HYPOCOTYLS 3, OBO1, ORGAN BOUNDARY 1 |
0.380 |
0.039 |
| 69 |
251227_at |
AT3G62700
|
ABCC14, ATP-binding cassette C14, ATMRP10, multidrug resistance-associated protein 10, MRP10, multidrug resistance-associated protein 10 |
0.370 |
0.007 |
| 70 |
258195_at |
AT3G13890
|
ATMYB26, myb domain protein 26, MS35, MALE STERILE 35, MYB26, myb domain protein 26 |
0.369 |
0.030 |
| 71 |
267035_at |
AT2G38400
|
AGT3, alanine:glyoxylate aminotransferase 3 |
0.369 |
-0.063 |
| 72 |
249068_at |
AT5G43980
|
PDLP1, plasmodesmata-located protein 1, PDLP1A, PLASMODESMATA-LOCATED PROTEIN 1A |
0.355 |
0.029 |
| 73 |
265277_at |
AT2G28410
|
unknown |
0.338 |
-0.002 |
| 74 |
263015_at |
AT1G23440
|
[Peptidase C15, pyroglutamyl peptidase I-like] |
0.336 |
-0.027 |
| 75 |
267300_at |
AT2G30140
|
UGT87A2, UDP-glucosyl transferase 87A2 |
0.335 |
-0.001 |
| 76 |
264611_at |
AT1G04680
|
[Pectin lyase-like superfamily protein] |
-0.994 |
-0.018 |
| 77 |
252691_at |
AT3G44050
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.963 |
0.239 |
| 78 |
248963_at |
AT5G45700
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.937 |
0.174 |
| 79 |
262081_at |
AT1G59540
|
ZCF125 |
-0.892 |
0.149 |
| 80 |
261460_at |
AT1G07880
|
ATMPK13 |
-0.875 |
-0.061 |
| 81 |
261198_at |
AT1G12940
|
ATNRT2.5, nitrate transporter2.5, NRT2.5, nitrate transporter2.5 |
-0.872 |
0.051 |
| 82 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
-0.868 |
0.012 |
| 83 |
258464_at |
AT3G17360
|
POK1, phragmoplast orienting kinesin 1 |
-0.832 |
0.035 |
| 84 |
252763_at |
AT3G42725
|
[Putative membrane lipoprotein] |
-0.827 |
0.012 |
| 85 |
255265_at |
AT4G05190
|
ATK5, kinesin 5 |
-0.821 |
0.139 |
| 86 |
248413_at |
AT5G51600
|
ATMAP65-3, ARABIDOPSIS THALIANA MICROTUBULE-ASSOCIATED PROTEIN 65-3, MAP65-3, MICROTUBULE-ASSOCIATED PROTEIN 65-3, PLE, PLEIADE |
-0.788 |
0.089 |
| 87 |
262494_at |
AT1G21810
|
unknown |
-0.786 |
0.182 |
| 88 |
259851_at |
AT1G72250
|
[Di-glucose binding protein with Kinesin motor domain] |
-0.768 |
0.028 |
| 89 |
257115_at |
AT3G20150
|
[Kinesin motor family protein] |
-0.766 |
0.033 |
| 90 |
259151_at |
AT3G10310
|
[P-loop nucleoside triphosphate hydrolases superfamily protein with CH (Calponin Homology) domain] |
-0.765 |
0.123 |
| 91 |
259930_at |
AT1G34355
|
ATPS1, PARALLEL SPINDLE 1, PS1, PARALLEL SPINDLE 1 |
-0.748 |
0.129 |
| 92 |
261780_at |
AT1G76310
|
CYCB2;4, CYCLIN B2;4 |
-0.748 |
0.109 |
| 93 |
248055_at |
AT5G55830
|
LecRK-S.7, L-type lectin receptor kinase S.7 |
-0.745 |
-0.003 |
| 94 |
249644_at |
AT5G37010
|
unknown |
-0.733 |
0.055 |
| 95 |
253978_at |
AT4G26660
|
unknown |
-0.732 |
0.123 |
| 96 |
247962_at |
AT5G56580
|
ANQ1, ARABIDOPSIS NQK1, ATMKK6, ARABIDOPSIS THALIANA MAP KINASE KINASE 6, MKK6, MAP kinase kinase 6 |
-0.703 |
0.075 |
| 97 |
267636_at |
AT2G42110
|
unknown |
-0.688 |
0.032 |
| 98 |
254379_at |
AT4G21820
|
[binding] |
-0.683 |
0.086 |
| 99 |
248476_at |
AT5G50890
|
[alpha/beta-Hydrolases superfamily protein] |
-0.683 |
0.051 |
| 100 |
256864_at |
AT3G23890
|
ATTOPII, TOPII, topoisomerase II |
-0.678 |
0.085 |
| 101 |
261840_at |
AT1G16070
|
AtTLP8, tubby like protein 8, TLP8, tubby like protein 8 |
-0.672 |
0.021 |
| 102 |
265085_at |
AT1G03780
|
AtTPX2, TPX2, targeting protein for XKLP2 |
-0.663 |
0.080 |
| 103 |
257978_at |
AT3G20860
|
ATNEK5, NIMA-related kinase 5, NEK5, NIMA-related kinase 5 |
-0.655 |
0.116 |
| 104 |
253148_at |
AT4G35620
|
CYCB2;2, Cyclin B2;2 |
-0.647 |
0.078 |
| 105 |
265349_at |
AT2G22610
|
[Di-glucose binding protein with Kinesin motor domain] |
-0.645 |
0.055 |
| 106 |
248691_at |
AT5G48310
|
unknown |
-0.624 |
-0.006 |
| 107 |
253757_at |
AT4G28950
|
ARAC7, Arabidopsis RAC-like 7, ATRAC7, ATROP9, RAC7, ROP9, RHO-related protein from plants 9 |
-0.613 |
0.066 |
| 108 |
251832_at |
AT3G55150
|
ATEXO70H1, exocyst subunit exo70 family protein H1, EXO70H1, exocyst subunit exo70 family protein H1 |
-0.605 |
0.065 |
| 109 |
248938_at |
AT5G45780
|
[Leucine-rich repeat protein kinase family protein] |
-0.605 |
-0.029 |
| 110 |
248658_at |
AT5G48600
|
ATCAP-C, ARABIDOPSIS THALIANA CHROMOSOME ASSOCIATED PROTEIN-C, ATSMC3, structural maintenance of chromosome 3, ATSMC4, ARABIDOPSIS THALIANA STRUCTURAL MAINTENANCE OF CHROMOSOME 4, SMC3, structural maintenance of chromosome 3 |
-0.593 |
0.035 |
| 111 |
251388_at |
AT3G60840
|
MAP65-4, microtubule-associated protein 65-4 |
-0.591 |
0.044 |
| 112 |
262109_at |
AT1G02730
|
ATCSLD5, cellulose synthase-like D5, CSLD5, CELLULOSE SYNTHASE LIKE D5, CSLD5, cellulose synthase-like D5, SOS6, SALT OVERLY SENSITIVE 6 |
-0.586 |
0.090 |
| 113 |
262036_at |
AT1G35530
|
At-FANCM, FANCM, Fanconi anemia complementation group M |
-0.585 |
0.101 |
| 114 |
245739_at |
AT1G44110
|
CYCA1;1, Cyclin A1;1 |
-0.578 |
0.118 |
| 115 |
258376_at |
AT3G17680
|
[Kinase interacting (KIP1-like) family protein] |
-0.576 |
0.055 |
| 116 |
248074_at |
AT5G55730
|
FLA1, FASCICLIN-like arabinogalactan 1 |
-0.576 |
0.001 |
| 117 |
262752_at |
AT1G16330
|
CYCB3;1, cyclin b3;1 |
-0.574 |
0.036 |
| 118 |
252089_at |
AT3G52110
|
unknown |
-0.571 |
0.054 |
| 119 |
264377_at |
AT2G25060
|
AtENODL14, ENODL14, early nodulin-like protein 14 |
-0.571 |
0.064 |
| 120 |
246920_at |
AT5G25090
|
AtENODL13, ENODL13, early nodulin-like protein 13 |
-0.545 |
0.111 |
| 121 |
246802_at |
AT5G27000
|
ATK4, kinesin 4, KATD, KINESIN-LIKE PROTEIN IN ARABIDOPSIS THALIANA D |
-0.545 |
-0.007 |
| 122 |
254118_at |
AT4G24790
|
[AAA-type ATPase family protein] |
-0.543 |
-0.007 |
| 123 |
252149_at |
AT3G51290
|
APSR1, Altered Phosphate Starvation Response 1 |
-0.541 |
0.091 |
| 124 |
250601_at |
AT5G07810
|
[SNF2 domain-containing protein / helicase domain-containing protein / HNH endonuclease domain-containing protein] |
-0.536 |
0.108 |
| 125 |
250504_at |
AT5G09840
|
[Putative endonuclease or glycosyl hydrolase] |
-0.532 |
0.044 |
| 126 |
258471_at |
AT3G06030
|
ANP3, NPK1-related protein kinase 3, AtANP3, MAPKKK12, NP3, NPK1-related protein kinase 3 |
-0.526 |
0.146 |
| 127 |
261364_at |
AT1G53140
|
DRP5A, Dynamin related protein 5A |
-0.522 |
0.066 |
| 128 |
247134_at |
AT5G66230
|
[Chalcone-flavanone isomerase family protein] |
-0.519 |
0.059 |
| 129 |
257524_at |
AT3G01330
|
DEL3, DP-E2F-like protein 3, E2FF, E2L2, E2F-LIKE 2 |
-0.517 |
-0.074 |
| 130 |
251527_at |
AT3G58650
|
TRM7, TON1 Recruiting Motif 7 |
-0.515 |
0.034 |
| 131 |
254808_at |
AT4G12740
|
[HhH-GPD base excision DNA repair family protein] |
-0.514 |
-0.071 |
| 132 |
266508_at |
AT2G47920
|
NET3C, Networked 3C |
-0.511 |
0.005 |
| 133 |
264895_at |
AT1G23100
|
[GroES-like family protein] |
-0.497 |
-0.018 |
| 134 |
258702_at |
AT3G09730
|
unknown |
-0.495 |
0.036 |
| 135 |
262336_at |
AT1G64220
|
TOM7-2, translocase of outer membrane 7 kDa subunit 2 |
-0.482 |
-0.012 |
| 136 |
259448_at |
AT1G13790
|
FDM4, factor of DNA methylation 4 |
-0.477 |
-0.026 |
| 137 |
246315_at |
AT3G56870
|
unknown |
-0.474 |
0.050 |
| 138 |
250196_at |
AT5G14580
|
[polyribonucleotide nucleotidyltransferase, putative] |
-0.474 |
-0.014 |
| 139 |
252877_at |
AT4G39630
|
unknown |
-0.468 |
-0.045 |
| 140 |
263441_at |
AT2G28620
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.461 |
0.046 |
| 141 |
249837_at |
AT5G23480
|
[SWIB/MDM2 domain] |
-0.452 |
-0.014 |
| 142 |
259978_at |
AT1G76540
|
CDKB2;1, cyclin-dependent kinase B2;1 |
-0.447 |
0.041 |
| 143 |
261085_at |
AT1G17480
|
IQD7, IQ-domain 7 |
-0.443 |
0.072 |
| 144 |
251035_at |
AT5G02220
|
SMR4, SIAMESE-RELATED 4 |
-0.441 |
-0.010 |
| 145 |
249032_at |
AT5G44910
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.437 |
-0.051 |
| 146 |
250499_at |
AT5G09730
|
ATBX3, ATBXL3, BETA-XYLOSIDASE 3, BX3, BXL3, beta-xylosidase 3, XYL3 |
-0.435 |
0.088 |
| 147 |
257005_at |
AT3G14190
|
CMR1, COPPER MODIFIED RESISTANCE 1 |
-0.433 |
0.072 |
| 148 |
261384_at |
AT1G05440
|
[C-8 sterol isomerases] |
-0.432 |
0.134 |
| 149 |
254443_at |
AT4G21070
|
ATBRCA1, ARABIDOPSIS THALIANA BREAST CANCER SUSCEPTIBILITY1, BRCA1, breast cancer susceptibility1 |
-0.415 |
0.053 |