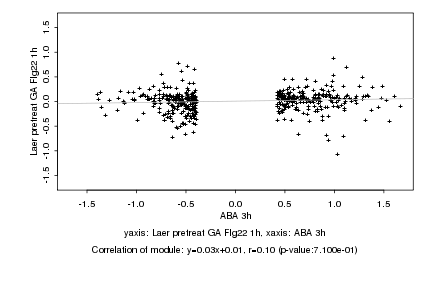
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ABA 3h |
Log2 signal ratio
Laer pretreat GA Flg22 1h |
| 1 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
1.664 |
-0.086 |
| 2 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.607 |
0.121 |
| 3 |
266415_at |
AT2G38530
|
cdf3, cell growth defect factor-3, LP2, LTP2, lipid transfer protein 2 |
1.550 |
-0.401 |
| 4 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
1.519 |
0.041 |
| 5 |
262448_at |
AT1G49450
|
[Transducin/WD40 repeat-like superfamily protein] |
1.484 |
0.323 |
| 6 |
265119_at |
AT1G62570
|
FMO GS-OX4, flavin-monooxygenase glucosinolate S-oxygenase 4 |
1.467 |
0.069 |
| 7 |
254697_at |
AT4G17970
|
ALMT12, aluminum-activated, malate transporter 12, ATALMT12, QUAC1, quick-activating anion channel 1 |
1.441 |
-0.103 |
| 8 |
254413_at |
AT4G21440
|
ATM4, A. THALIANA MYB 4, ATMYB102, MYB-like 102, MYB102, MYB102, MYB-like 102 |
1.385 |
0.297 |
| 9 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
1.369 |
-0.180 |
| 10 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
1.342 |
0.111 |
| 11 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
1.329 |
0.129 |
| 12 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
1.311 |
0.118 |
| 13 |
252073_at |
AT3G51750
|
unknown |
1.306 |
-0.367 |
| 14 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.291 |
0.120 |
| 15 |
251824_at |
AT3G55090
|
ABCG16, ATP-binding cassette G16 |
1.279 |
0.502 |
| 16 |
266503_at |
AT2G47780
|
[Rubber elongation factor protein (REF)] |
1.277 |
0.056 |
| 17 |
248227_at |
AT5G53820
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.251 |
0.314 |
| 18 |
263385_at |
AT2G40170
|
ATEM6, ARABIDOPSIS EARLY METHIONINE-LABELLED 6, EM6, EARLY METHIONINE-LABELLED 6, GEA6, LATE EMBRYOGENESIS ABUNDANT 6 |
1.219 |
-0.034 |
| 19 |
250500_at |
AT5G09530
|
PELPK1, Pro-Glu-Leu|Ile|Val-Pro-Lys 1, PRP10, proline-rich protein 10 |
1.214 |
0.113 |
| 20 |
249289_at |
AT5G41040
|
ASFT, aliphatic suberin feruloyl-transferase, HHT, hydroxycinnamoyl- CoA:ω-hydroxyacid O-hydroxycinnamoyltransferase, RWP1, REDUCED LEVELS OF WALL-BOUND PHENOLICS 1 |
1.213 |
0.031 |
| 21 |
260357_at |
AT1G69260
|
AFP1, ABI five binding protein |
1.213 |
0.077 |
| 22 |
252019_at |
AT3G53040
|
[late embryogenesis abundant protein, putative / LEA protein, putative] |
1.168 |
0.018 |
| 23 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
1.167 |
0.046 |
| 24 |
249771_at |
AT5G24080
|
[Protein kinase superfamily protein] |
1.147 |
0.066 |
| 25 |
249941_at |
AT5G22270
|
unknown |
1.124 |
0.140 |
| 26 |
256073_at |
AT1G18100
|
E12A11, MFT, MOTHER OF FT AND TFL1 |
1.121 |
0.120 |
| 27 |
256937_at |
AT3G22620
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.115 |
0.691 |
| 28 |
247657_at |
AT5G59845
|
[Gibberellin-regulated family protein] |
1.111 |
0.005 |
| 29 |
264436_at |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
1.102 |
-0.022 |
| 30 |
258224_at |
AT3G15670
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.102 |
0.072 |
| 31 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
1.100 |
-0.166 |
| 32 |
265411_at |
AT2G16630
|
[Pollen Ole e 1 allergen and extensin family protein] |
1.092 |
-0.123 |
| 33 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
1.088 |
-0.708 |
| 34 |
248209_at |
AT5G53990
|
[UDP-Glycosyltransferase superfamily protein] |
1.088 |
0.309 |
| 35 |
266462_at |
AT2G47770
|
ATTSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related, TSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related |
1.069 |
-0.074 |
| 36 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
1.051 |
0.081 |
| 37 |
263495_at |
AT2G42530
|
COR15B, cold regulated 15b |
1.040 |
-0.121 |
| 38 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
1.037 |
0.030 |
| 39 |
247902_at |
AT5G57350
|
AHA3, H(+)-ATPase 3, ATAHA3, ARABIDOPSIS THALIANA ARABIDOPSIS H(+)-ATPASE, HA3, H(+)-ATPase 3 |
1.026 |
-0.099 |
| 40 |
266421_at |
AT2G38540
|
ATLTP1, ARABIDOPSIS THALIANA LIPID TRANSFER PROTEIN 1, LP1, lipid transfer protein 1, LTP1, LIPID TRANSFER PROTEIN 1 |
1.023 |
-1.056 |
| 41 |
245412_at |
AT4G17280
|
[Auxin-responsive family protein] |
1.022 |
0.134 |
| 42 |
256022_at |
AT1G58360
|
AAP1, amino acid permease 1, AtAAP1, NAT2, NEUTRAL AMINO ACID TRANSPORTER 2 |
1.015 |
-0.015 |
| 43 |
264729_at |
AT1G22990
|
HIPP22, heavy metal associated isoprenylated plant protein 22 |
1.013 |
0.095 |
| 44 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.997 |
0.075 |
| 45 |
248352_at |
AT5G52300
|
LTI65, LOW-TEMPERATURE-INDUCED 65, RD29B, RESPONSIVE TO DESSICATION 29B |
0.988 |
-0.085 |
| 46 |
256933_at |
AT3G22600
|
LTPG5, glycosylphosphatidylinositol-anchored lipid protein transfer 5 |
0.988 |
0.545 |
| 47 |
258957_at |
AT3G01420
|
ALPHA-DOX1, alpha-dioxygenase 1, DIOX1, DOX1, PADOX-1, plant alpha dioxygenase 1 |
0.984 |
0.885 |
| 48 |
267559_at |
AT2G45570
|
CYP76C2, cytochrome P450, family 76, subfamily C, polypeptide 2 |
0.983 |
0.228 |
| 49 |
266900_at |
AT2G34610
|
unknown |
0.978 |
-0.015 |
| 50 |
265024_at |
AT1G24600
|
unknown |
0.977 |
0.408 |
| 51 |
248505_at |
AT5G50360
|
unknown |
0.976 |
0.095 |
| 52 |
263998_at |
AT2G22510
|
[hydroxyproline-rich glycoprotein family protein] |
0.972 |
0.098 |
| 53 |
245386_at |
AT4G14010
|
RALFL32, ralf-like 32 |
0.964 |
0.038 |
| 54 |
248333_at |
AT5G52390
|
[PAR1 protein] |
0.961 |
0.312 |
| 55 |
245748_at |
AT1G51140
|
AKS1, ABA-responsive kinase substrate 1, FBH3, FLOWERING BHLH 3 |
0.956 |
0.099 |
| 56 |
259380_at |
AT3G16340
|
ABCG29, ATP-binding cassette G29, AtABCG29, ATPDR1, PLEIOTROPIC DRUG RESISTANCE 1, PDR1, pleiotropic drug resistance 1 |
0.953 |
0.036 |
| 57 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
0.947 |
0.162 |
| 58 |
247463_at |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
0.943 |
0.134 |
| 59 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
0.934 |
-0.289 |
| 60 |
259592_at |
AT1G27950
|
LTPG1, glycosylphosphatidylinositol-anchored lipid protein transfer 1 |
0.934 |
-0.786 |
| 61 |
256378_at |
AT1G66830
|
[Leucine-rich repeat protein kinase family protein] |
0.933 |
0.206 |
| 62 |
253619_at |
AT4G30460
|
[glycine-rich protein] |
0.930 |
-0.107 |
| 63 |
260040_at |
AT1G68765
|
IDA, INFLORESCENCE DEFICIENT IN ABSCISSION |
0.919 |
0.155 |
| 64 |
258751_at |
AT3G05890
|
RCI2B, RARE-COLD-INDUCIBLE 2B |
0.919 |
0.118 |
| 65 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
0.913 |
0.326 |
| 66 |
263709_at |
AT1G09310
|
unknown |
0.911 |
-0.678 |
| 67 |
264102_at |
AT1G79270
|
ECT8, evolutionarily conserved C-terminal region 8 |
0.909 |
-0.103 |
| 68 |
247421_at |
AT5G62800
|
[Protein with RING/U-box and TRAF-like domains] |
0.903 |
0.106 |
| 69 |
263073_at |
AT2G17500
|
[Auxin efflux carrier family protein] |
0.888 |
0.227 |
| 70 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.887 |
0.103 |
| 71 |
255723_at |
AT3G29575
|
AFP3, ABI five binding protein 3 |
0.881 |
0.155 |
| 72 |
254667_at |
AT4G18280
|
[glycine-rich cell wall protein-related] |
0.876 |
-0.301 |
| 73 |
256757_at |
AT3G25620
|
ABCG21, ATP-binding cassette G21 |
0.875 |
0.130 |
| 74 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
0.875 |
-0.367 |
| 75 |
254201_at |
AT4G24130
|
unknown |
0.863 |
0.262 |
| 76 |
255877_at |
AT2G40460
|
[Major facilitator superfamily protein] |
0.860 |
-0.142 |
| 77 |
261570_at |
AT1G01120
|
KCS1, 3-ketoacyl-CoA synthase 1 |
0.858 |
-0.100 |
| 78 |
265188_at |
AT1G23800
|
ALDH2B, aldehyde dehydrogenase 2B, ALDH2B7, aldehyde dehydrogenase 2B7 |
0.854 |
0.007 |
| 79 |
260264_at |
AT1G68500
|
unknown |
0.847 |
0.080 |
| 80 |
256114_at |
AT1G16850
|
unknown |
0.840 |
0.156 |
| 81 |
251668_at |
AT3G57010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.838 |
0.031 |
| 82 |
247957_at |
AT5G57050
|
ABI2, ABA INSENSITIVE 2, AtABI2 |
0.835 |
0.054 |
| 83 |
266143_at |
AT2G38905
|
[Low temperature and salt responsive protein family] |
0.834 |
-0.035 |
| 84 |
259573_at |
AT1G20390
|
[gypsy-like retrotransposon family, has a 1.5e-251 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
0.833 |
0.082 |
| 85 |
245982_at |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
0.833 |
0.153 |
| 86 |
253293_at |
AT4G33905
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
0.833 |
0.067 |
| 87 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
0.832 |
0.070 |
| 88 |
246582_at |
AT1G31750
|
[proline-rich family protein] |
0.828 |
-0.148 |
| 89 |
265511_at |
AT2G05540
|
[Glycine-rich protein family] |
0.821 |
-0.183 |
| 90 |
246922_at |
AT5G25110
|
CIPK25, CBL-interacting protein kinase 25, SnRK3.25, SNF1-RELATED PROTEIN KINASE 3.25 |
0.815 |
0.040 |
| 91 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
0.808 |
0.070 |
| 92 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
0.807 |
0.129 |
| 93 |
254287_at |
AT4G22960
|
unknown |
0.806 |
0.413 |
| 94 |
249063_at |
AT5G44110
|
ABCI21, ATP-binding cassette A21, ATNAP2, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN, ATPOP1, POP1 |
0.801 |
-0.188 |
| 95 |
265913_at |
AT2G25625
|
unknown |
0.800 |
0.143 |
| 96 |
265216_at |
AT1G05100
|
MAPKKK18, mitogen-activated protein kinase kinase kinase 18 |
0.797 |
0.058 |
| 97 |
253875_at |
AT4G27520
|
AtENODL2, ENODL2, early nodulin-like protein 2 |
0.796 |
-0.081 |
| 98 |
254226_at |
AT4G23690
|
AtDIR6, Arabidopsis thaliana dirigent protein 6, DIR6, dirigent protein 6 |
0.786 |
0.230 |
| 99 |
252004_at |
AT3G52780
|
ATPAP20, PAP20 |
0.782 |
0.093 |
| 100 |
258139_at |
AT3G24520
|
AT-HSFC1, HSFC1, heat shock transcription factor C1 |
0.779 |
-0.018 |
| 101 |
264217_at |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
0.770 |
0.036 |
| 102 |
251658_at |
AT3G57020
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.749 |
-0.018 |
| 103 |
252691_at |
AT3G44050
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.745 |
0.017 |
| 104 |
252863_at |
AT4G39800
|
ATIPS1, INOSITOL 3-PHOSPHATE SYNTHASE 1, ATMIPS1, MYO-INOSITOL-1-PHOSPHATE SYNTHASE 1, MI-1-P SYNTHASE, MIPS1, D-myo-Inositol 3-Phosphate Synthase 1, MIPS1, myo-inositol-1-phosphate synthase 1 |
0.744 |
-0.389 |
| 105 |
244986_at |
ATCG00820
|
RPS19, ribosomal protein S19 |
0.734 |
-0.090 |
| 106 |
265475_at |
AT2G15620
|
ATHNIR, ARABIDOPSIS THALIANA NITRITE REDUCTASE, NIR, NITRITE REDUCTASE, NIR1, nitrite reductase 1 |
0.733 |
-0.154 |
| 107 |
265122_at |
AT1G62540
|
FMO GS-OX2, flavin-monooxygenase glucosinolate S-oxygenase 2 |
0.731 |
0.044 |
| 108 |
258752_at |
AT3G09520
|
ATEXO70H4, exocyst subunit exo70 family protein H4, EXO70H4, exocyst subunit exo70 family protein H4 |
0.724 |
0.312 |
| 109 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.721 |
-0.257 |
| 110 |
265276_at |
AT2G28400
|
unknown |
0.717 |
0.023 |
| 111 |
263492_at |
AT2G42560
|
[late embryogenesis abundant domain-containing protein / LEA domain-containing protein] |
0.717 |
0.047 |
| 112 |
256069_at |
AT1G13740
|
AFP2, ABI five binding protein 2 |
0.715 |
0.080 |
| 113 |
258397_at |
AT3G15357
|
unknown |
0.715 |
-0.011 |
| 114 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
0.711 |
0.453 |
| 115 |
247487_at |
AT5G62150
|
[peptidoglycan-binding LysM domain-containing protein] |
0.701 |
0.300 |
| 116 |
255546_at |
AT4G01910
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.683 |
0.100 |
| 117 |
251566_at |
AT3G58210
|
[TRAF-like family protein] |
0.682 |
0.077 |
| 118 |
266001_at |
AT2G24150
|
HHP3, heptahelical protein 3 |
0.680 |
-0.232 |
| 119 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.680 |
0.166 |
| 120 |
250624_at |
AT5G07330
|
unknown |
0.679 |
0.103 |
| 121 |
262236_at |
AT1G48330
|
unknown |
0.678 |
-0.015 |
| 122 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.673 |
0.126 |
| 123 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.670 |
0.166 |
| 124 |
267496_at |
AT2G30550
|
DALL3, DAD1-Like Lipase 3 |
0.669 |
0.211 |
| 125 |
267080_at |
AT2G41190
|
[Transmembrane amino acid transporter family protein] |
0.662 |
0.059 |
| 126 |
250158_at |
AT5G15190
|
unknown |
0.654 |
0.108 |
| 127 |
250252_at |
AT5G13750
|
ZIFL1, zinc induced facilitator-like 1 |
0.654 |
0.026 |
| 128 |
262164_at |
AT1G78070
|
[Transducin/WD40 repeat-like superfamily protein] |
0.651 |
-0.006 |
| 129 |
266655_at |
AT2G25880
|
AtAUR2, ataurora2, AUR2, ataurora2 |
0.650 |
0.061 |
| 130 |
261511_at |
AT1G71770
|
PAB5, poly(A)-binding protein 5 |
0.639 |
0.201 |
| 131 |
259430_at |
AT1G01610
|
ATGPAT4, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 4, GPAT4, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 4 |
0.635 |
-0.655 |
| 132 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
0.634 |
-0.172 |
| 133 |
245112_at |
AT2G41540
|
GPDHC1 |
0.629 |
0.301 |
| 134 |
253203_at |
AT4G34710
|
ADC2, arginine decarboxylase 2, ATADC2, SPE2 |
0.621 |
0.099 |
| 135 |
245724_at |
AT1G73390
|
[Endosomal targeting BRO1-like domain-containing protein] |
0.614 |
-0.116 |
| 136 |
263238_at |
AT2G16580
|
SAUR8, SMALL AUXIN UPREGULATED RNA 8 |
0.613 |
0.070 |
| 137 |
256674_at |
AT3G52360
|
unknown |
0.612 |
-0.024 |
| 138 |
262933_at |
AT1G65840
|
ATPAO4, polyamine oxidase 4, PAO4, polyamine oxidase 4 |
0.612 |
0.067 |
| 139 |
254998_at |
AT4G09760
|
[Protein kinase superfamily protein] |
0.609 |
-0.155 |
| 140 |
255959_at |
AT1G21980
|
ATPIP5K1, phosphatidylinositol-4-phosphate 5-kinase 1, ATPIPK1, PIP5K1, phosphatidylinositol-4-phosphate 5-kinase 1 |
0.603 |
-0.161 |
| 141 |
257642_at |
AT3G25710
|
ATAIG1, BHLH32, basic helix-loop-helix 32, TMO5, TARGET OF MONOPTEROS 5 |
0.594 |
0.115 |
| 142 |
259661_at |
AT1G55265
|
unknown |
0.592 |
0.052 |
| 143 |
249411_at |
AT5G40390
|
RS5, raffinose synthase 5, SIP1, seed imbibition 1-like |
0.589 |
0.067 |
| 144 |
252119_at |
AT3G51030
|
ATTRX H1, ARABIDOPSIS THALIANA THIOREDOXIN H-TYPE 1, ATTRX1, thioredoxin H-type 1, TRX1, thioredoxin H-type 1 |
0.587 |
0.001 |
| 145 |
249465_at |
AT5G39720
|
AIG2L, avirulence induced gene 2 like protein |
0.585 |
0.248 |
| 146 |
253994_at |
AT4G26080
|
ABI1, ABA INSENSITIVE 1, AtABI1 |
0.581 |
0.141 |
| 147 |
256964_at |
AT3G13520
|
AGP12, arabinogalactan protein 12, ATAGP12 |
0.579 |
0.103 |
| 148 |
258167_at |
AT3G21560
|
BRT1, Bright Trichomes 1, UGT84A2, UDP-glucosyl transferase 84A2 |
0.576 |
-0.001 |
| 149 |
260560_at |
AT2G43590
|
[Chitinase family protein] |
0.576 |
0.036 |
| 150 |
263825_at |
AT2G40370
|
LAC5, laccase 5 |
0.572 |
0.067 |
| 151 |
266489_at |
AT2G35190
|
ATNPSN11, NPSN11, novel plant snare 11, NSPN11 |
0.571 |
0.452 |
| 152 |
248100_at |
AT5G55180
|
[O-Glycosyl hydrolases family 17 protein] |
0.568 |
-0.062 |
| 153 |
245629_at |
AT1G56580
|
SVB, SMALLER WITH VARIABLE BRANCHES |
0.566 |
0.028 |
| 154 |
250690_at |
AT5G06530
|
ABCG22, ATP-binding cassette G22, AtABCG22, Arabidopsis thaliana ATP-binding cassette G22 |
0.565 |
-0.365 |
| 155 |
256120_at |
AT1G18130
|
[Class II aaRS and biotin synthetases superfamily protein] |
0.555 |
0.101 |
| 156 |
255028_at |
AT4G09890
|
unknown |
0.552 |
0.190 |
| 157 |
253066_at |
AT4G37770
|
ACS8, 1-amino-cyclopropane-1-carboxylate synthase 8 |
0.546 |
0.291 |
| 158 |
256759_at |
AT3G25640
|
unknown |
0.543 |
0.011 |
| 159 |
252872_at |
AT4G40010
|
SNRK2-7, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-7, SNRK2.7, SNF1-related protein kinase 2.7, SRK2F |
0.543 |
0.066 |
| 160 |
267008_at |
AT2G39350
|
ABCG1, ATP-binding cassette G1 |
0.541 |
0.108 |
| 161 |
259466_at |
AT1G19050
|
ARR7, response regulator 7 |
0.539 |
0.073 |
| 162 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
0.536 |
0.148 |
| 163 |
249794_at |
AT5G23530
|
AtCXE18, carboxyesterase 18, CXE18, carboxyesterase 18 |
0.534 |
0.029 |
| 164 |
262912_at |
AT1G59740
|
AtNPF4.3, NPF4.3, NRT1/ PTR family 4.3 |
0.533 |
0.053 |
| 165 |
249284_at |
AT5G41810
|
unknown |
0.532 |
-0.109 |
| 166 |
248896_at |
AT5G46350
|
ATWRKY8, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 8, WRKY8, WRKY DNA-binding protein 8 |
0.529 |
0.182 |
| 167 |
247097_at |
AT5G66460
|
AtMAN7, MAN7, endo-beta-mannase 7 |
0.529 |
-0.083 |
| 168 |
255479_at |
AT4G02380
|
AtLEA5, Arabidopsis thaliana late embryogenensis abundant like 5, SAG21, senescence-associated gene 21 |
0.529 |
0.070 |
| 169 |
259845_at |
AT1G73590
|
ATPIN1, ARABIDOPSIS THALIANA PIN-FORMED 1, PIN1, PIN-FORMED 1 |
0.526 |
0.017 |
| 170 |
258901_at |
AT3G05640
|
[Protein phosphatase 2C family protein] |
0.526 |
-0.038 |
| 171 |
254562_at |
AT4G19230
|
CYP707A1, cytochrome P450, family 707, subfamily A, polypeptide 1 |
0.524 |
0.258 |
| 172 |
260853_at |
AT1G21950
|
unknown |
0.523 |
0.061 |
| 173 |
247134_at |
AT5G66230
|
[Chalcone-flavanone isomerase family protein] |
0.521 |
0.163 |
| 174 |
263574_at |
AT2G16990
|
[Major facilitator superfamily protein] |
0.518 |
0.081 |
| 175 |
247470_at |
AT5G62220
|
ATGT18, glycosyltransferase 18, GT18, glycosyltransferase 18, XLT2, Xyloglucan L-side chain galactosylTransferase position 2 |
0.512 |
-0.042 |
| 176 |
252408_at |
AT3G47600
|
ATMYB94, myb domain protein 94, ATMYBCP70, MYB94, myb domain protein 94 |
0.502 |
-0.116 |
| 177 |
254917_at |
AT4G11350
|
unknown |
0.500 |
0.075 |
| 178 |
254085_at |
AT4G24960
|
ATHVA22D, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE D, HVA22D, HVA22 homologue D |
0.493 |
-0.144 |
| 179 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
0.492 |
-0.101 |
| 180 |
253582_at |
AT4G30670
|
[Putative membrane lipoprotein] |
0.492 |
0.456 |
| 181 |
250160_at |
AT5G15210
|
ATHB30, homeobox protein 30, HB30, homeobox protein 30, ZFHD3, ZINC FINGER HOMEODOMAIN 3, ZHD8, ZINC FINGER HOMEODOMAIN 8 |
0.491 |
0.110 |
| 182 |
263016_at |
AT1G23410
|
[Ribosomal protein S27a / Ubiquitin family protein] |
0.490 |
0.049 |
| 183 |
246097_at |
AT5G20270
|
HHP1, heptahelical transmembrane protein1 |
0.490 |
-0.350 |
| 184 |
246994_at |
AT5G67460
|
[O-Glycosyl hydrolases family 17 protein] |
0.490 |
0.140 |
| 185 |
245784_at |
AT1G32190
|
[alpha/beta-Hydrolases superfamily protein] |
0.487 |
0.033 |
| 186 |
261315_at |
AT1G53170
|
ATERF-8, ATERF8, ETHYLENE RESPONSE ELEMENT BINDING FACTOR 4, ERF8, ethylene response factor 8 |
0.486 |
-0.079 |
| 187 |
266705_at |
AT2G19750
|
[Ribosomal protein S30 family protein] |
0.485 |
0.051 |
| 188 |
252133_at |
AT3G50900
|
unknown |
0.482 |
0.122 |
| 189 |
254326_at |
AT4G22610
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.478 |
0.059 |
| 190 |
259516_at |
AT1G20450
|
ERD10, EARLY RESPONSIVE TO DEHYDRATION 10, LTI29, LOW TEMPERATURE INDUCED 29, LTI45, LOW TEMPERATURE INDUCED 45 |
0.476 |
-0.067 |
| 191 |
260885_at |
AT1G29230
|
ATCIPK18, ATWL1, WPL4-LIKE 1, CIPK18, CBL-interacting protein kinase 18, SnRK3.20, SNF1-RELATED PROTEIN KINASE 3.20, WL1, WPL4-LIKE 1 |
0.476 |
0.230 |
| 192 |
248413_at |
AT5G51600
|
ATMAP65-3, ARABIDOPSIS THALIANA MICROTUBULE-ASSOCIATED PROTEIN 65-3, MAP65-3, MICROTUBULE-ASSOCIATED PROTEIN 65-3, PLE, PLEIADE |
0.476 |
0.095 |
| 193 |
252010_at |
AT3G52740
|
unknown |
0.474 |
-0.180 |
| 194 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.470 |
0.167 |
| 195 |
264200_at |
AT1G22650
|
A/N-InvD, alkaline/neutral invertase D |
0.470 |
-0.220 |
| 196 |
262436_at |
AT1G47610
|
[Transducin/WD40 repeat-like superfamily protein] |
0.469 |
0.140 |
| 197 |
245486_x_at |
AT4G16240
|
unknown |
0.468 |
-0.051 |
| 198 |
258275_at |
AT3G15760
|
unknown |
0.467 |
0.067 |
| 199 |
261876_at |
AT1G50590
|
[RmlC-like cupins superfamily protein] |
0.466 |
-0.057 |
| 200 |
263796_at |
AT2G24540
|
AFR, ATTENUATED FAR-RED RESPONSE |
0.463 |
-0.195 |
| 201 |
254633_at |
AT4G18640
|
MRH1, morphogenesis of root hair 1 |
0.461 |
0.082 |
| 202 |
247056_at |
AT5G66750
|
ATDDM1, CHA1, CHR01, CHROMATIN REMODELING 1, CHR1, chromatin remodeling 1, DDM1, DECREASED DNA METHYLATION 1, SOM1, SOMNIFEROUS 1, SOM4 |
0.460 |
0.023 |
| 203 |
266009_at |
AT2G37420
|
[ATP binding microtubule motor family protein] |
0.460 |
0.021 |
| 204 |
246781_at |
AT5G27350
|
SFP1 |
0.458 |
0.139 |
| 205 |
258732_at |
AT3G05820
|
A/N-InvH, alkaline/neutral invertase H, At-A/N-InvH, Arabidopsis alkaline/neutral invertase H, INVH, invertase H |
0.458 |
0.109 |
| 206 |
249910_at |
AT5G22630
|
ADT5, arogenate dehydratase 5 |
0.452 |
0.205 |
| 207 |
260010_at |
AT1G68020
|
ATTPS6, TPS6, TREHALOSE -6-PHOSPHATASE SYNTHASE S6 |
0.446 |
-0.046 |
| 208 |
261363_at |
AT1G41830
|
SKS6, SKU5 SIMILAR 6, SKS6, SKU5-similar 6 |
0.445 |
-0.232 |
| 209 |
246025_at |
AT5G21150
|
AGO9, ARGONAUTE 9 |
0.443 |
0.106 |
| 210 |
245904_at |
AT5G11110
|
ATSPS2F, sucrose phosphate synthase 2F, KNS2, KAONASHI 2, SPS1, SUCROSE PHOSPHATE SYNTHASE 1, SPS2F, sucrose phosphate synthase 2F, SPSA2, sucrose-phosphate synthase A2 |
0.442 |
-0.130 |
| 211 |
250892_at |
AT5G03760
|
ATCSLA09, ATCSLA9, CSLA09, CSLA9, CELLULOSE SYNTHASE LIKE A9, RAT4, RESISTANT TO AGROBACTERIUM TRANSFORMATION 4 |
0.442 |
-0.002 |
| 212 |
250493_at |
AT5G09800
|
[ARM repeat superfamily protein] |
0.441 |
0.193 |
| 213 |
248685_at |
AT5G48500
|
unknown |
0.440 |
0.215 |
| 214 |
248132_at |
AT5G54840
|
ATSGP1, SGP1 |
0.438 |
0.157 |
| 215 |
256736_at |
AT3G29410
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
0.436 |
0.175 |
| 216 |
264252_at |
AT1G09180
|
ATSAR1, SECRETION-ASSOCIATED RAS 1, ATSARA1A, secretion-associated RAS super family 1, SARA1A, secretion-associated RAS super family 1 |
0.434 |
0.124 |
| 217 |
257766_at |
AT3G23030
|
IAA2, indole-3-acetic acid inducible 2 |
0.432 |
0.044 |
| 218 |
254372_at |
AT4G21620
|
[glycine-rich protein] |
0.426 |
-0.183 |
| 219 |
244983_at |
ATCG00790
|
RPL16, ribosomal protein L16 |
0.425 |
-0.099 |
| 220 |
247706_at |
AT5G59480
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.425 |
-0.047 |
| 221 |
261754_at |
AT1G76130
|
AMY2, alpha-amylase-like 2, ATAMY2, ARABIDOPSIS THALIANA ALPHA-AMYLASE-LIKE 2 |
0.424 |
-0.081 |
| 222 |
253978_at |
AT4G26660
|
unknown |
0.421 |
0.067 |
| 223 |
251238_at |
AT3G62430
|
[Protein with RNI-like/FBD-like domains] |
0.421 |
0.192 |
| 224 |
263867_at |
AT2G36830
|
GAMMA-TIP, gamma tonoplast intrinsic protein, GAMMA-TIP1, GAMMA TONOPLAST INTRINSIC PROTEIN 1, TIP1;1, TONOPLAST INTRINSIC PROTEIN 1;1 |
0.420 |
-0.366 |
| 225 |
263128_at |
AT1G78600
|
BBX22, B-box domain protein 22, DBB3, DOUBLE B-BOX 3, LZF1, light-regulated zinc finger protein 1, STH3, SALT TOLERANCE HOMOLOG 3 |
0.419 |
0.135 |
| 226 |
260668_at |
AT1G19530
|
unknown |
-1.397 |
0.158 |
| 227 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
-1.388 |
0.052 |
| 228 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
-1.369 |
0.203 |
| 229 |
263851_at |
AT2G04460
|
unknown |
-1.356 |
-0.114 |
| 230 |
266118_at |
AT2G02130
|
LCR68, low-molecular-weight cysteine-rich 68, PDF2.3 |
-1.319 |
-0.277 |
| 231 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
-1.278 |
0.034 |
| 232 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
-1.201 |
-0.162 |
| 233 |
246001_at |
AT5G20790
|
unknown |
-1.186 |
0.072 |
| 234 |
260741_at |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
-1.165 |
0.206 |
| 235 |
259399_at |
AT1G17710
|
AtPEPC1, Arabidopsis thaliana phosphoethanolamine/phosphocholine phosphatase 1, PEPC1, phosphoethanolamine/phosphocholine phosphatase 1 |
-1.136 |
0.007 |
| 236 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
-1.130 |
-0.016 |
| 237 |
262284_at |
AT1G68670
|
[myb-like transcription factor family protein] |
-1.126 |
-0.032 |
| 238 |
249151_at |
AT5G43360
|
ATPT4, PHT1;3, phosphate transporter 1;3, PHT3, PHOSPHATE TRANSPORTER 3 |
-1.081 |
0.184 |
| 239 |
265387_at |
AT2G20670
|
unknown |
-1.045 |
0.046 |
| 240 |
254001_at |
AT4G26260
|
MIOX4, myo-inositol oxygenase 4 |
-1.036 |
0.185 |
| 241 |
259694_at |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
-1.027 |
0.061 |
| 242 |
250091_at |
AT5G17340
|
[Putative membrane lipoprotein] |
-1.021 |
0.027 |
| 243 |
255016_at |
AT4G10120
|
ATSPS4F |
-0.991 |
-0.371 |
| 244 |
252234_at |
AT3G49780
|
ATPSK3 (FORMER SYMBOL), ATPSK4, phytosulfokine 4 precursor, PSK4, phytosulfokine 4 precursor |
-0.975 |
0.282 |
| 245 |
253087_at |
AT4G36350
|
ATPAP25, ARABIDOPSIS THALIANA PURPLE ACID PHOSPHATASE 25, PAP25, purple acid phosphatase 25 |
-0.965 |
0.112 |
| 246 |
256787_at |
AT3G13790
|
ATBFRUCT1, ATCWINV1, ARABIDOPSIS THALIANA CELL WALL INVERTASE 1 |
-0.943 |
0.123 |
| 247 |
265116_at |
AT1G62480
|
[Vacuolar calcium-binding protein-related] |
-0.934 |
0.149 |
| 248 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-0.933 |
-0.241 |
| 249 |
249527_at |
AT5G38710
|
[Methylenetetrahydrofolate reductase family protein] |
-0.915 |
0.121 |
| 250 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
-0.890 |
0.122 |
| 251 |
258856_at |
AT3G02040
|
AtGDPD1, GDPD1, Glycerophosphodiester phosphodiesterase 1, SRG3, senescence-related gene 3 |
-0.890 |
0.043 |
| 252 |
246215_at |
AT4G37180
|
[Homeodomain-like superfamily protein] |
-0.887 |
0.049 |
| 253 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
-0.882 |
0.051 |
| 254 |
246072_at |
AT5G20240
|
PI, PISTILLATA |
-0.876 |
0.262 |
| 255 |
248931_at |
AT5G46040
|
[Major facilitator superfamily protein] |
-0.873 |
0.045 |
| 256 |
245528_at |
AT4G15530
|
PPDK, pyruvate orthophosphate dikinase |
-0.853 |
0.062 |
| 257 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.836 |
-0.095 |
| 258 |
246855_at |
AT5G26280
|
[TRAF-like family protein] |
-0.835 |
0.317 |
| 259 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
-0.829 |
0.007 |
| 260 |
252176_at |
AT3G50720
|
[Protein kinase superfamily protein] |
-0.828 |
0.075 |
| 261 |
258227_at |
AT3G15620
|
UVR3, UV REPAIR DEFECTIVE 3 |
-0.825 |
0.003 |
| 262 |
246963_at |
AT5G24820
|
[Eukaryotic aspartyl protease family protein] |
-0.823 |
0.064 |
| 263 |
245026_at |
ATCG00140
|
ATPH |
-0.822 |
-0.021 |
| 264 |
254797_at |
AT4G13030
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.817 |
-0.034 |
| 265 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
-0.815 |
0.132 |
| 266 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
-0.776 |
-0.204 |
| 267 |
257331_at |
ATMG01330
|
ORF107H |
-0.773 |
-0.122 |
| 268 |
262366_at |
AT1G72890
|
[Disease resistance protein (TIR-NBS class)] |
-0.772 |
0.237 |
| 269 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
-0.772 |
0.031 |
| 270 |
250059_at |
AT5G17820
|
[Peroxidase superfamily protein] |
-0.770 |
0.077 |
| 271 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
-0.768 |
0.158 |
| 272 |
252387_at |
AT3G47800
|
[Galactose mutarotase-like superfamily protein] |
-0.767 |
-0.021 |
| 273 |
253322_at |
AT4G33980
|
unknown |
-0.756 |
0.559 |
| 274 |
250940_at |
AT5G03310
|
SAUR44, SMALL AUXIN UPREGULATED RNA 44 |
-0.739 |
0.062 |
| 275 |
258225_at |
AT3G15630
|
unknown |
-0.730 |
0.147 |
| 276 |
252167_at |
AT3G50560
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.730 |
-0.265 |
| 277 |
266884_at |
AT2G44790
|
UCC2, uclacyanin 2 |
-0.728 |
0.368 |
| 278 |
248411_at |
AT5G51580
|
unknown |
-0.728 |
0.106 |
| 279 |
252183_at |
AT3G50740
|
UGT72E1, UDP-glucosyl transferase 72E1 |
-0.722 |
-0.374 |
| 280 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
-0.721 |
0.291 |
| 281 |
263391_at |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
-0.720 |
-0.385 |
| 282 |
256989_at |
AT3G28580
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.717 |
0.292 |
| 283 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
-0.713 |
-0.260 |
| 284 |
246888_at |
AT5G26270
|
unknown |
-0.701 |
0.126 |
| 285 |
258570_at |
AT3G04530
|
ATPPCK2, PEPCK2, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 2, PPCK2, phosphoenolpyruvate carboxylase kinase 2 |
-0.698 |
0.052 |
| 286 |
260411_at |
AT1G69890
|
unknown |
-0.693 |
0.074 |
| 287 |
262947_at |
AT1G75750
|
GASA1, GAST1 protein homolog 1 |
-0.693 |
-0.312 |
| 288 |
267628_at |
AT2G42280
|
AKS3, ABA-responsive kinase substrate 3, FBH4, FLOWERING BHLH 4 |
-0.690 |
0.104 |
| 289 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
-0.688 |
0.299 |
| 290 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
-0.686 |
0.034 |
| 291 |
249010_at |
AT5G44580
|
unknown |
-0.681 |
-0.196 |
| 292 |
257798_at |
AT3G15950
|
NAI2 |
-0.680 |
-0.241 |
| 293 |
260662_at |
AT1G19540
|
[NmrA-like negative transcriptional regulator family protein] |
-0.679 |
-0.037 |
| 294 |
252220_at |
AT3G49940
|
LBD38, LOB domain-containing protein 38 |
-0.671 |
-0.017 |
| 295 |
251509_at |
AT3G59010
|
PME35, pectin methylesterase 35, PME61, pectin methylesterase 61 |
-0.670 |
0.045 |
| 296 |
247954_at |
AT5G56870
|
BGAL4, beta-galactosidase 4 |
-0.669 |
0.140 |
| 297 |
262525_at |
AT1G17060
|
CHI2, CHIBI 2, CYP72C1, cytochrome p450 72c1, SHK1, SHRINK 1, SOB7, SUPPRESSOR OF PHYB-4 7 |
-0.660 |
0.285 |
| 298 |
248703_at |
AT5G48430
|
[Eukaryotic aspartyl protease family protein] |
-0.660 |
0.140 |
| 299 |
246075_at |
AT5G20410
|
ATMGD2, ARABIDOPSIS THALIANA MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE 2, MGD2, monogalactosyldiacylglycerol synthase 2 |
-0.658 |
-0.326 |
| 300 |
258270_at |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
-0.657 |
-0.297 |
| 301 |
255543_at |
AT4G01870
|
[tolB protein-related] |
-0.649 |
0.082 |
| 302 |
248167_at |
AT5G54530
|
unknown |
-0.647 |
-0.071 |
| 303 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
-0.646 |
-0.242 |
| 304 |
258055_at |
AT3G16250
|
NDF4, NDH-dependent cyclic electron flow 1, PnsB3, Photosynthetic NDH subcomplex B 3 |
-0.640 |
-0.724 |
| 305 |
260431_at |
AT1G68190
|
BBX27, B-box domain protein 27 |
-0.640 |
-0.180 |
| 306 |
263133_at |
AT1G78450
|
[SOUL heme-binding family protein] |
-0.639 |
-0.405 |
| 307 |
255852_at |
AT1G66970
|
GDPDL1, Glycerophosphodiester phosphodiesterase (GDPD) like 1, SVL2, SHV3-like 2 |
-0.637 |
-0.069 |
| 308 |
253332_at |
AT4G33420
|
[Peroxidase superfamily protein] |
-0.635 |
0.116 |
| 309 |
263352_at |
AT2G22080
|
unknown |
-0.634 |
0.059 |
| 310 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
-0.633 |
0.032 |
| 311 |
258537_at |
AT3G04210
|
[Disease resistance protein (TIR-NBS class)] |
-0.632 |
-0.225 |
| 312 |
261658_at |
AT1G50040
|
unknown |
-0.630 |
-0.095 |
| 313 |
247946_at |
AT5G57180
|
CIA2, chloroplast import apparatus 2 |
-0.626 |
-0.113 |
| 314 |
251576_at |
AT3G58200
|
[TRAF-like family protein] |
-0.625 |
0.060 |
| 315 |
261518_at |
AT1G71695
|
[Peroxidase superfamily protein] |
-0.617 |
-0.137 |
| 316 |
255360_at |
AT4G03960
|
AtPFA-DSP4, PFA-DSP4, plant and fungi atypical dual-specificity phosphatase 4 |
-0.615 |
-0.002 |
| 317 |
261814_at |
AT1G08310
|
[alpha/beta-Hydrolases superfamily protein] |
-0.605 |
0.278 |
| 318 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
-0.603 |
0.039 |
| 319 |
251491_at |
AT3G59480
|
[pfkB-like carbohydrate kinase family protein] |
-0.602 |
0.116 |
| 320 |
255734_at |
AT1G25550
|
[myb-like transcription factor family protein] |
-0.600 |
0.095 |
| 321 |
262830_at |
AT1G14700
|
ATPAP3, PAP3, purple acid phosphatase 3 |
-0.599 |
-0.512 |
| 322 |
255491_at |
AT4G02670
|
AtIDD12, indeterminate(ID)-domain 12, IDD12, indeterminate(ID)-domain 12 |
-0.598 |
0.027 |
| 323 |
249729_at |
AT5G24410
|
PGL4, 6-phosphogluconolactonase 4 |
-0.596 |
0.081 |
| 324 |
255433_at |
AT4G03210
|
XTH9, xyloglucan endotransglucosylase/hydrolase 9 |
-0.594 |
-0.087 |
| 325 |
259161_at |
AT3G01500
|
ATBCA1, BETA CARBONIC ANHYDRASE 1, ATSABP3, ARABIDOPSIS THALIANA SALICYLIC ACID-BINDING PROTEIN 3, CA1, carbonic anhydrase 1, SABP3, SALICYLIC ACID-BINDING PROTEIN 3 |
-0.594 |
-0.156 |
| 326 |
250032_at |
AT5G18170
|
GDH1, glutamate dehydrogenase 1 |
-0.593 |
0.125 |
| 327 |
264247_at |
AT1G60160
|
[Potassium transporter family protein] |
-0.591 |
-0.542 |
| 328 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
-0.589 |
0.151 |
| 329 |
260969_at |
AT1G12240
|
ATBETAFRUCT4, AtFRUCT4, AtVI2, FRUCT4, fructosidase 4, VAC-INV, VACUOLAR INVERTASE, VI2, vacuolar invertase 2 |
-0.585 |
-0.034 |
| 330 |
266712_at |
AT2G46750
|
AtGulLO2, GulLO2, L -gulono-1,4-lactone ( L -GulL) oxidase 2 |
-0.585 |
0.776 |
| 331 |
266782_at |
AT2G29120
|
ATGLR2.7, glutamate receptor 2.7, GLR2.7, GLUTAMATE RECEPTOR 2.7, GLR2.7, glutamate receptor 2.7 |
-0.582 |
-0.098 |
| 332 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
-0.581 |
0.113 |
| 333 |
254280_at |
AT4G22756
|
ATSMO1-2, SMO1-2, sterol C4-methyl oxidase 1-2 |
-0.578 |
-0.084 |
| 334 |
251144_at |
AT5G01210
|
[HXXXD-type acyl-transferase family protein] |
-0.575 |
0.056 |
| 335 |
254111_at |
AT4G24890
|
ATPAP24, ARABIDOPSIS THALIANA PURPLE ACID PHOSPHATASE 24, PAP24, purple acid phosphatase 24 |
-0.574 |
0.070 |
| 336 |
260201_at |
AT1G67600
|
[Acid phosphatase/vanadium-dependent haloperoxidase-related protein] |
-0.571 |
0.152 |
| 337 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
-0.567 |
0.044 |
| 338 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.566 |
0.071 |
| 339 |
244960_at |
ATCG01020
|
RPL32, ribosomal protein L32 |
-0.565 |
0.029 |
| 340 |
258541_at |
AT3G07000
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.563 |
0.012 |
| 341 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
-0.559 |
-0.488 |
| 342 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
-0.551 |
-0.054 |
| 343 |
264501_at |
AT1G09390
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.551 |
-0.228 |
| 344 |
247072_at |
AT5G66490
|
unknown |
-0.550 |
-0.012 |
| 345 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
-0.548 |
-0.026 |
| 346 |
249035_at |
AT5G44190
|
ATGLK2, GLK2, GOLDEN2-like 2, GPRI2, GBF'S PRO-RICH REGION-INTERACTING FACTOR 2 |
-0.548 |
-0.074 |
| 347 |
256181_at |
AT1G51820
|
[Leucine-rich repeat protein kinase family protein] |
-0.546 |
0.106 |
| 348 |
266976_at |
AT2G39410
|
[alpha/beta-Hydrolases superfamily protein] |
-0.546 |
0.622 |
| 349 |
264893_at |
AT1G23140
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.546 |
-0.005 |
| 350 |
263217_at |
AT1G30740
|
[FAD-binding Berberine family protein] |
-0.543 |
0.429 |
| 351 |
252411_at |
AT3G47430
|
PEX11B, peroxin 11B |
-0.540 |
-0.442 |
| 352 |
259358_at |
AT1G13250
|
GATL3, galacturonosyltransferase-like 3 |
-0.539 |
0.116 |
| 353 |
260422_at |
AT1G69630
|
[F-box/RNI-like superfamily protein] |
-0.536 |
0.059 |
| 354 |
252199_at |
AT3G50270
|
[HXXXD-type acyl-transferase family protein] |
-0.531 |
-0.434 |
| 355 |
260429_at |
AT1G72450
|
JAZ6, jasmonate-zim-domain protein 6, TIFY11B, TIFY DOMAIN PROTEIN 11B |
-0.531 |
0.015 |
| 356 |
245262_at |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
-0.530 |
-0.462 |
| 357 |
260032_at |
AT1G68750
|
ATPPC4, phosphoenolpyruvate carboxylase 4, PPC4, phosphoenolpyruvate carboxylase 4 |
-0.530 |
-0.010 |
| 358 |
260296_at |
AT1G63750
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.529 |
-0.029 |
| 359 |
261662_at |
AT1G18350
|
ATMKK7, MAP kinase kinase 7, BUD1, BUSHY AND DWARF 1, MKK7, MAP kinase kinase 7, MKK7, MAP KINASE KINASE7 |
-0.528 |
-0.184 |
| 360 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
-0.519 |
-0.055 |
| 361 |
253653_at |
AT4G30050
|
unknown |
-0.514 |
0.063 |
| 362 |
256304_at |
AT1G69523
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.513 |
-0.297 |
| 363 |
260388_at |
AT1G74070
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.509 |
-0.451 |
| 364 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
-0.509 |
-0.094 |
| 365 |
256255_at |
AT3G11280
|
[Duplicated homeodomain-like superfamily protein] |
-0.508 |
-0.296 |
| 366 |
262288_at |
AT1G70760
|
CRR23, CHLORORESPIRATORY REDUCTION 23, NdhL, NADH dehydrogenase-like complex L |
-0.506 |
-0.660 |
| 367 |
251032_at |
AT5G02030
|
BLH9, BEL1-LIKE HOMEODOMAIN 9, BLR, BELLRINGER, HB-6, LSN, LARSON, PNY, PENNYWISE, RPL, REPLUMLESS, VAN, VAAMANA |
-0.505 |
-0.135 |
| 368 |
252160_at |
AT3G50570
|
[hydroxyproline-rich glycoprotein family protein] |
-0.503 |
0.063 |
| 369 |
246949_at |
AT5G25140
|
CYP71B13, cytochrome P450, family 71, subfamily B, polypeptide 13 |
-0.503 |
-0.327 |
| 370 |
264700_at |
AT1G70100
|
unknown |
-0.502 |
-0.126 |
| 371 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
-0.500 |
0.185 |
| 372 |
264561_at |
AT1G55810
|
UKL3, uridine kinase-like 3 |
-0.495 |
-0.061 |
| 373 |
246195_at |
AT4G36410
|
UBC17, ubiquitin-conjugating enzyme 17 |
-0.495 |
0.170 |
| 374 |
255854_at |
AT1G67050
|
unknown |
-0.493 |
0.272 |
| 375 |
256332_at |
AT1G76890
|
AT-GT2, GT2 |
-0.493 |
0.054 |
| 376 |
247625_at |
AT5G60200
|
TMO6, TARGET OF MONOPTEROS 6 |
-0.492 |
0.010 |
| 377 |
259316_at |
AT3G01175
|
unknown |
-0.491 |
0.714 |
| 378 |
256965_at |
AT3G13450
|
DIN4, DARK INDUCIBLE 4 |
-0.491 |
0.126 |
| 379 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
-0.487 |
0.145 |
| 380 |
260317_at |
AT1G63800
|
UBC5, ubiquitin-conjugating enzyme 5 |
-0.485 |
-0.135 |
| 381 |
260783_at |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
-0.485 |
-0.281 |
| 382 |
247128_at |
AT5G66110
|
HIPP27, heavy metal associated isoprenylated plant protein 27 |
-0.483 |
-0.090 |
| 383 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
-0.481 |
0.088 |
| 384 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
-0.478 |
0.227 |
| 385 |
256168_at |
AT1G51805
|
[Leucine-rich repeat protein kinase family protein] |
-0.477 |
-0.228 |
| 386 |
249790_at |
AT5G24290
|
MEB2, MEMBRANE OF ER BODY 2 |
-0.477 |
0.026 |
| 387 |
254013_at |
AT4G26050
|
PIRL8, plant intracellular ras group-related LRR 8 |
-0.471 |
0.370 |
| 388 |
249824_at |
AT5G23380
|
unknown |
-0.470 |
0.063 |
| 389 |
251983_at |
AT3G53210
|
UMAMIT6, Usually multiple acids move in and out Transporters 6 |
-0.466 |
-0.143 |
| 390 |
254408_at |
AT4G21390
|
B120 |
-0.465 |
0.027 |
| 391 |
251143_at |
AT5G01220
|
SQD2, sulfoquinovosyldiacylglycerol 2 |
-0.465 |
-0.391 |
| 392 |
245130_at |
AT2G45340
|
[Leucine-rich repeat protein kinase family protein] |
-0.460 |
-0.289 |
| 393 |
247985_at |
AT5G56790
|
[Protein kinase superfamily protein] |
-0.458 |
-0.044 |
| 394 |
257655_at |
AT3G13350
|
[HMG (high mobility group) box protein with ARID/BRIGHT DNA-binding domain] |
-0.458 |
0.018 |
| 395 |
260855_at |
AT1G21920
|
[Histone H3 K4-specific methyltransferase SET7/9 family protein] |
-0.458 |
-0.175 |
| 396 |
258223_at |
AT3G15840
|
PIFI, post-illumination chlorophyll fluorescence increase |
-0.453 |
0.036 |
| 397 |
263106_at |
AT2G05160
|
[CCCH-type zinc fingerfamily protein with RNA-binding domain] |
-0.451 |
0.157 |
| 398 |
266617_at |
AT2G29670
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.451 |
0.037 |
| 399 |
246114_at |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
-0.450 |
0.072 |
| 400 |
258025_at |
AT3G19480
|
3-PGDH, PGDH3, phosphoglycerate dehydrogenase 3 |
-0.449 |
-0.331 |
| 401 |
249045_at |
AT5G44380
|
[FAD-binding Berberine family protein] |
-0.446 |
0.112 |
| 402 |
258990_at |
AT3G08840
|
[D-alanine--D-alanine ligase family] |
-0.444 |
-0.147 |
| 403 |
258189_at |
AT3G17860
|
JAI3, JASMONATE-INSENSITIVE 3, JAZ3, jasmonate-zim-domain protein 3, TIFY6B |
-0.443 |
-0.044 |
| 404 |
247352_at |
AT5G63650
|
SNRK2-5, SNRK2.5, SNF1-related protein kinase 2.5, SRK2H, SNF1-RELATED PROTEIN KINASE 2H |
-0.443 |
0.105 |
| 405 |
251132_at |
AT5G01200
|
[Duplicated homeodomain-like superfamily protein] |
-0.443 |
0.183 |
| 406 |
266355_at |
AT2G01400
|
unknown |
-0.442 |
-0.095 |
| 407 |
250022_at |
AT5G18210
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.441 |
0.004 |
| 408 |
248058_at |
AT5G55530
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.441 |
0.087 |
| 409 |
253980_at |
AT4G26620
|
[Sucrase/ferredoxin-like family protein] |
-0.439 |
-0.134 |
| 410 |
263052_at |
AT2G13430
|
unknown |
-0.439 |
0.047 |
| 411 |
264987_at |
AT1G27030
|
unknown |
-0.434 |
-0.231 |
| 412 |
265472_at |
AT2G15580
|
[RING/U-box superfamily protein] |
-0.433 |
-0.289 |
| 413 |
246098_at |
AT5G20400
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.432 |
0.065 |
| 414 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
-0.432 |
0.012 |
| 415 |
256596_at |
AT3G28540
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.432 |
-0.010 |
| 416 |
261762_at |
AT1G15510
|
ATECB2, ARABIDOPSIS EARLY CHLOROPLAST BIOGENESIS2, ECB2, EARLY CHLOROPLAST BIOGENESIS2, VAC1, VANILLA CREAM 1 |
-0.431 |
-0.434 |
| 417 |
255608_at |
AT4G01140
|
unknown |
-0.431 |
0.028 |
| 418 |
257946_at |
AT3G21710
|
unknown |
-0.430 |
0.378 |
| 419 |
247440_at |
AT5G62680
|
AtNPF2.11, GTR2, GLUCOSINOLATE TRANSPORTER-2, NPF2.11, NRT1/ PTR family 2.11 |
-0.428 |
0.031 |
| 420 |
258355_at |
AT3G14330
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.428 |
-0.025 |
| 421 |
256570_at |
AT3G19540
|
unknown |
-0.427 |
0.054 |
| 422 |
250398_at |
AT5G11000
|
unknown |
-0.425 |
0.115 |
| 423 |
248050_at |
AT5G56100
|
[glycine-rich protein / oleosin] |
-0.425 |
-0.038 |
| 424 |
255719_at |
AT1G32080
|
AtLrgB, LrgB |
-0.425 |
-0.267 |
| 425 |
252543_at |
AT3G45780
|
JK224, NPH1, NONPHOTOTROPIC HYPOCOTYL 1, PHOT1, phototropin 1, RPT1, ROOT PHOTOTROPISM 1 |
-0.424 |
-0.621 |
| 426 |
252698_at |
AT3G43670
|
[Copper amine oxidase family protein] |
-0.424 |
0.136 |
| 427 |
247326_at |
AT5G64110
|
[Peroxidase superfamily protein] |
-0.422 |
0.660 |
| 428 |
247696_at |
AT5G59780
|
ATMYB59, MYB DOMAIN PROTEIN 59, ATMYB59-1, ATMYB59-2, ATMYB59-3, MYB59, myb domain protein 59 |
-0.421 |
-0.295 |
| 429 |
259211_at |
AT3G09020
|
[alpha 1,4-glycosyltransferase family protein] |
-0.420 |
0.190 |
| 430 |
260884_at |
AT1G29240
|
unknown |
-0.418 |
-0.110 |
| 431 |
252991_at |
AT4G38470
|
STY46, serine/threonine/tyrosine kinase 46 |
-0.418 |
0.081 |
| 432 |
258299_at |
AT3G23410
|
ATFAO3, ARABIDOPSIS FATTY ALCOHOL OXIDASE 3, FAO3, fatty alcohol oxidase 3 |
-0.416 |
-0.341 |
| 433 |
265481_at |
AT2G15960
|
unknown |
-0.415 |
0.072 |
| 434 |
255463_at |
AT4G02960
|
ATRE2, retro element 2, RE2, retro element 2 |
-0.415 |
-0.015 |
| 435 |
267077_at |
AT2G40970
|
MYBC1 |
-0.415 |
-0.167 |
| 436 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.412 |
0.230 |
| 437 |
262629_at |
AT1G06460
|
ACD31.2, ALPHA-CRYSTALLIN DOMAIN 31.2, ACD32.1, alpha-crystallin domain 32.1 |
-0.412 |
-0.141 |
| 438 |
263126_at |
AT1G78460
|
[SOUL heme-binding family protein] |
-0.408 |
-0.250 |
| 439 |
267462_at |
AT2G33735
|
[Chaperone DnaJ-domain superfamily protein] |
-0.408 |
-0.200 |
| 440 |
258932_at |
AT3G10150
|
ATPAP16, PAP16, purple acid phosphatase 16 |
-0.407 |
-0.037 |
| 441 |
257773_at |
AT3G29185
|
unknown |
-0.407 |
-0.452 |
| 442 |
259379_at |
AT3G16350
|
[Homeodomain-like superfamily protein] |
-0.406 |
-0.218 |
| 443 |
249693_at |
AT5G35750
|
AHK2, histidine kinase 2, HK2, histidine kinase 2 |
-0.406 |
0.103 |
| 444 |
246028_at |
AT5G21170
|
5'-AMP-activated protein kinase beta-2 subunit protein |
-0.406 |
0.050 |
| 445 |
247924_at |
AT5G57655
|
[xylose isomerase family protein] |
-0.405 |
0.098 |
| 446 |
264889_at |
AT1G23050
|
hydroxyproline-rich glycoprotein family protein |
-0.405 |
-0.076 |
| 447 |
261211_at |
AT1G12780
|
ATUGE1, A. THALIANA UDP-GLC 4-EPIMERASE 1, UGE1, UDP-D-glucose/UDP-D-galactose 4-epimerase 1 |
-0.402 |
0.041 |
| 448 |
264956_at |
AT1G76990
|
ACR3, ACT domain repeat 3 |
-0.402 |
-0.218 |
| 449 |
262958_at |
AT1G54410
|
AtHIRD11, HIRD11, dehydrin 11kDa |
-0.402 |
-0.015 |
| 450 |
256772_at |
AT3G13750
|
BGAL1, beta galactosidase 1, BGAL1, beta-galactosidase 1 |
-0.401 |
-0.350 |