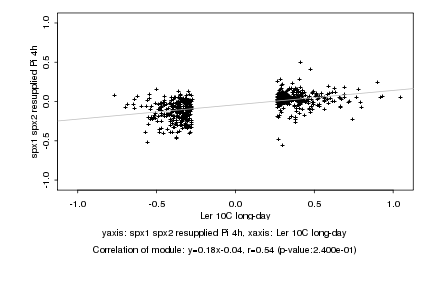
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Ler 10C long-day |
Log2 signal ratio
spx1 spx2 resupplied Pi 4h |
| 1 |
251531_at |
AT3G58550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.044 |
0.059 |
| 2 |
265334_at |
AT2G18370
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.930 |
0.071 |
| 3 |
254956_at |
AT4G10850
|
AtSWEET7, SWEET7 |
0.915 |
0.058 |
| 4 |
254098_at |
AT4G25100
|
ATFSD1, ARABIDOPSIS FE SUPEROXIDE DISMUTASE 1, FSD1, Fe superoxide dismutase 1 |
0.899 |
0.254 |
| 5 |
249101_at |
AT5G43580
|
UPI, UNUSUAL SERINE PROTEASE INHIBITOR |
0.796 |
-0.069 |
| 6 |
247729_at |
AT5G59530
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.788 |
-0.005 |
| 7 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
0.777 |
0.163 |
| 8 |
262307_at |
AT1G71000
|
[Chaperone DnaJ-domain superfamily protein] |
0.763 |
0.056 |
| 9 |
248959_at |
AT5G45630
|
unknown |
0.742 |
-0.220 |
| 10 |
245889_at |
AT5G09480
|
[hydroxyproline-rich glycoprotein family protein] |
0.722 |
0.005 |
| 11 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
0.716 |
-0.006 |
| 12 |
247657_at |
AT5G59845
|
[Gibberellin-regulated family protein] |
0.689 |
0.191 |
| 13 |
249556_at |
AT5G38195
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.687 |
0.055 |
| 14 |
251188_at |
AT3G62730
|
unknown |
0.686 |
0.094 |
| 15 |
267389_at |
AT2G44460
|
BGLU28, beta glucosidase 28 |
0.671 |
0.030 |
| 16 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
0.661 |
-0.061 |
| 17 |
250500_at |
AT5G09530
|
PELPK1, Pro-Glu-Leu|Ile|Val-Pro-Lys 1, PRP10, proline-rich protein 10 |
0.661 |
-0.071 |
| 18 |
248194_at |
AT5G54095
|
unknown |
0.660 |
0.043 |
| 19 |
247236_at |
AT5G64590
|
unknown |
0.659 |
0.062 |
| 20 |
256743_at |
AT3G29370
|
P1R3, P1R3 |
0.632 |
0.122 |
| 21 |
260220_at |
AT1G74650
|
ATMYB31, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 31, ATY13, MYB31, myb domain protein 31 |
0.628 |
0.006 |
| 22 |
262454_at |
AT1G11190
|
BFN1, bifunctional nuclease i, ENDO1, ENDONUCLEASE 1 |
0.627 |
0.173 |
| 23 |
246889_at |
AT5G25470
|
[AP2/B3-like transcriptional factor family protein] |
0.615 |
0.008 |
| 24 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.612 |
0.115 |
| 25 |
257509_at |
AT1G63190
|
[Cystatin/monellin superfamily protein] |
0.599 |
0.045 |
| 26 |
248060_at |
AT5G55560
|
[Protein kinase superfamily protein] |
0.595 |
0.107 |
| 27 |
265962_at |
AT2G37460
|
UMAMIT12, Usually multiple acids move in and out Transporters 12 |
0.588 |
-0.016 |
| 28 |
264729_at |
AT1G22990
|
HIPP22, heavy metal associated isoprenylated plant protein 22 |
0.584 |
0.192 |
| 29 |
255302_at |
AT4G04830
|
ATMSRB5, methionine sulfoxide reductase B5, MSRB5, methionine sulfoxide reductase B5 |
0.583 |
0.092 |
| 30 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
0.580 |
0.036 |
| 31 |
249289_at |
AT5G41040
|
ASFT, aliphatic suberin feruloyl-transferase, HHT, hydroxycinnamoyl- CoA:ω-hydroxyacid O-hydroxycinnamoyltransferase, RWP1, REDUCED LEVELS OF WALL-BOUND PHENOLICS 1 |
0.563 |
-0.092 |
| 32 |
266918_at |
AT2G45800
|
PLIM2a, PLIM2a |
0.562 |
0.001 |
| 33 |
247324_at |
AT5G64190
|
unknown |
0.561 |
0.046 |
| 34 |
263144_at |
AT1G54070
|
[Dormancy/auxin associated family protein] |
0.560 |
0.063 |
| 35 |
258360_at |
AT3G14250
|
[RING/U-box superfamily protein] |
0.559 |
0.018 |
| 36 |
254430_at |
AT4G20820
|
[FAD-binding Berberine family protein] |
0.543 |
0.068 |
| 37 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
0.541 |
0.104 |
| 38 |
246861_at |
AT5G25890
|
IAA28, indole-3-acetic acid inducible 28, IAR2, IAA-ALANINE RESISTANT 2 |
0.531 |
-0.030 |
| 39 |
247210_at |
AT5G65020
|
ANNAT2, annexin 2, AtANN2 |
0.530 |
-0.054 |
| 40 |
257049_at |
AT3G15250
|
unknown |
0.527 |
0.069 |
| 41 |
260437_at |
AT1G68380
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
0.527 |
0.075 |
| 42 |
259124_at |
AT3G02310
|
AGL4, AGAMOUS-like 4, SEP2, SEPALLATA 2 |
0.526 |
0.045 |
| 43 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
0.519 |
-0.039 |
| 44 |
265341_at |
AT2G18360
|
[alpha/beta-Hydrolases superfamily protein] |
0.518 |
0.033 |
| 45 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.511 |
0.093 |
| 46 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.508 |
0.107 |
| 47 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
0.508 |
0.087 |
| 48 |
255811_at |
AT4G10250
|
ATHSP22.0 |
0.503 |
0.072 |
| 49 |
260101_at |
AT1G73260
|
ATKTI1, ARABIDOPSIS THALIANA KUNITZ TRYPSIN INHIBITOR 1, KTI1, kunitz trypsin inhibitor 1 |
0.493 |
0.129 |
| 50 |
265685_at |
AT2G24430
|
ANAC038, NAC domain containing protein 38, ANAC039, Arabidopsis NAC domain containing protein 39, NAC038, NAC domain containing protein 38 |
0.493 |
-0.011 |
| 51 |
254024_at |
AT4G25780
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.491 |
-0.032 |
| 52 |
264590_at |
AT2G17710
|
unknown |
0.488 |
0.112 |
| 53 |
245697_at |
AT5G04200
|
AtMC9, metacaspase 9, AtMCP2f, metacaspase 2f, MC9, metacaspase 9, MCP2f, metacaspase 2f |
0.488 |
0.118 |
| 54 |
251896_at |
AT3G54390
|
[sequence-specific DNA binding transcription factors] |
0.485 |
-0.003 |
| 55 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.483 |
-0.095 |
| 56 |
251827_at |
AT3G55120
|
A11, AtCHI, CFI, CHALCONE FLAVANONE ISOMERASE, CHI, chalcone isomerase, TT5, TRANSPARENT TESTA 5 |
0.479 |
-0.054 |
| 57 |
248911_at |
AT5G45830
|
ATDOG1, DOG1, DELAY OF GERMINATION 1, GAAS5, germination ability after storage 5, GSQ5, GLUCOSE SENSING QTL 5 |
0.476 |
0.037 |
| 58 |
263959_at |
AT2G36210
|
SAUR45, SMALL AUXIN UPREGULATED RNA 45 |
0.475 |
0.412 |
| 59 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
0.471 |
-0.128 |
| 60 |
254629_at |
AT4G18425
|
unknown |
0.468 |
0.073 |
| 61 |
260035_at |
AT1G68850
|
[Peroxidase superfamily protein] |
0.465 |
-0.078 |
| 62 |
253999_at |
AT4G26200
|
ACCS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ACS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ATACS7 |
0.464 |
-0.101 |
| 63 |
260560_at |
AT2G43590
|
[Chitinase family protein] |
0.460 |
-0.020 |
| 64 |
255659_at |
AT4G00895
|
[ATPase, F1 complex, OSCP/delta subunit protein] |
0.457 |
0.032 |
| 65 |
248735_at |
AT5G48100
|
ATLAC15, AtTT10, LAC15, LACCASE-LIKE 15, TT10, TRANSPARENT TESTA 10 |
0.457 |
0.001 |
| 66 |
256574_at |
AT3G14780
|
unknown |
0.454 |
0.044 |
| 67 |
248753_at |
AT5G47630
|
mtACP3, mitochondrial acyl carrier protein 3 |
0.449 |
0.024 |
| 68 |
265863_at |
AT2G01770
|
ATVIT1, VIT1, vacuolar iron transporter 1 |
0.440 |
-0.000 |
| 69 |
253884_at |
AT4G27670
|
HSP21, heat shock protein 21 |
0.438 |
0.077 |
| 70 |
251181_at |
AT3G62820
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.437 |
0.123 |
| 71 |
252081_at |
AT3G51910
|
AT-HSFA7A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A7A, HSFA7A, heat shock transcription factor A7A |
0.433 |
-0.177 |
| 72 |
263693_at |
AT1G31200
|
ATPP2-A9, phloem protein 2-A9, PP2-A9, phloem protein 2-A9 |
0.433 |
0.019 |
| 73 |
255575_at |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
0.429 |
0.034 |
| 74 |
248968_at |
AT5G45280
|
[Pectinacetylesterase family protein] |
0.425 |
0.005 |
| 75 |
262438_at |
AT1G47410
|
unknown |
0.424 |
-0.097 |
| 76 |
252751_at |
AT3G43430
|
[RING/U-box superfamily protein] |
0.420 |
0.104 |
| 77 |
250258_at |
AT5G13790
|
AGL15, AGAMOUS-like 15 |
0.419 |
0.071 |
| 78 |
256062_at |
AT1G07090
|
LSH6, LIGHT SENSITIVE HYPOCOTYLS 6 |
0.419 |
0.145 |
| 79 |
248100_at |
AT5G55180
|
[O-Glycosyl hydrolases family 17 protein] |
0.416 |
-0.009 |
| 80 |
266764_at |
AT2G47050
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.416 |
0.003 |
| 81 |
246183_at |
AT5G20940
|
[Glycosyl hydrolase family protein] |
0.416 |
0.028 |
| 82 |
262608_at |
AT1G14120
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.416 |
0.096 |
| 83 |
254627_at |
AT4G18395
|
unknown |
0.415 |
0.029 |
| 84 |
245792_at |
AT1G32100
|
ATPRR1, PRR1, pinoresinol reductase 1 |
0.415 |
-0.005 |
| 85 |
263475_at |
AT2G31945
|
unknown |
0.412 |
0.119 |
| 86 |
252561_at |
AT3G45980
|
H2B, HISTONE H2B, HTB9 |
0.411 |
-0.033 |
| 87 |
257821_at |
AT3G25170
|
RALFL26, ralf-like 26 |
0.410 |
0.034 |
| 88 |
254746_at |
AT4G12980
|
[Auxin-responsive family protein] |
0.410 |
0.090 |
| 89 |
258395_at |
AT3G15500
|
ANAC055, NAC domain containing protein 55, ATNAC3, NAC domain containing protein 3, NAC055, NAC domain containing protein 55, NAC3, NAC domain containing protein 3 |
0.408 |
0.183 |
| 90 |
254085_at |
AT4G24960
|
ATHVA22D, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE D, HVA22D, HVA22 homologue D |
0.407 |
-0.013 |
| 91 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
0.407 |
0.503 |
| 92 |
267388_at |
AT2G44450
|
BGLU15, beta glucosidase 15 |
0.406 |
0.006 |
| 93 |
262886_at |
AT1G14760
|
KNATM, KNOX Arabidopsis thaliana meinox |
0.406 |
0.064 |
| 94 |
263919_at |
AT2G36470
|
unknown |
0.404 |
-0.036 |
| 95 |
256741_at |
AT3G29375
|
[XH domain-containing protein] |
0.404 |
-0.088 |
| 96 |
266078_at |
AT2G40670
|
ARR16, response regulator 16, RR16, response regulator 16 |
0.401 |
-0.147 |
| 97 |
262411_at |
AT1G34640
|
[peptidases] |
0.401 |
0.291 |
| 98 |
258879_at |
AT3G03270
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.400 |
0.047 |
| 99 |
248685_at |
AT5G48500
|
unknown |
0.400 |
0.008 |
| 100 |
248118_at |
AT5G55050
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.399 |
0.067 |
| 101 |
248224_at |
AT5G53490
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.396 |
0.006 |
| 102 |
265722_at |
AT2G40100
|
LHCB4.3, light harvesting complex photosystem II |
0.396 |
0.125 |
| 103 |
257946_at |
AT3G21710
|
unknown |
0.396 |
0.140 |
| 104 |
251739_at |
AT3G56170
|
CAN, Ca-2+ dependent nuclease, CAN1, calcium dependent nuclease 1 |
0.395 |
0.005 |
| 105 |
246346_at |
AT3G56810
|
unknown |
0.394 |
-0.042 |
| 106 |
265188_at |
AT1G23800
|
ALDH2B, aldehyde dehydrogenase 2B, ALDH2B7, aldehyde dehydrogenase 2B7 |
0.394 |
-0.009 |
| 107 |
265118_at |
AT1G62660
|
[Glycosyl hydrolases family 32 protein] |
0.393 |
0.011 |
| 108 |
266755_at |
AT2G47150
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.392 |
0.043 |
| 109 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.392 |
0.081 |
| 110 |
249721_at |
AT5G24655
|
LSU4, RESPONSE TO LOW SULFUR 4 |
0.389 |
0.163 |
| 111 |
255806_at |
AT4G10260
|
[pfkB-like carbohydrate kinase family protein] |
0.389 |
0.052 |
| 112 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
0.389 |
-0.044 |
| 113 |
257151_at |
AT3G27200
|
[Cupredoxin superfamily protein] |
0.386 |
0.125 |
| 114 |
261749_at |
AT1G76180
|
ERD14, EARLY RESPONSE TO DEHYDRATION 14 |
0.385 |
-0.084 |
| 115 |
250597_at |
AT5G07680
|
ANAC079, Arabidopsis NAC domain containing protein 79, ANAC080, NAC domain containing protein 80, ATNAC4, NAC080, NAC domain containing protein 80, NAC4 |
0.385 |
0.036 |
| 116 |
257819_at |
AT3G25165
|
RALFL25, ralf-like 25 |
0.383 |
0.044 |
| 117 |
248491_at |
AT5G51010
|
[Rubredoxin-like superfamily protein] |
0.383 |
0.034 |
| 118 |
260549_at |
AT2G43535
|
[Scorpion toxin-like knottin superfamily protein] |
0.383 |
0.038 |
| 119 |
267079_at |
AT2G41200
|
unknown |
0.382 |
0.096 |
| 120 |
248545_at |
AT5G50260
|
CEP1, cysteine endopeptidase 1 |
0.381 |
-0.126 |
| 121 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
0.381 |
0.154 |
| 122 |
256883_at |
AT3G26440
|
unknown |
0.380 |
-0.193 |
| 123 |
254104_at |
AT4G25040
|
[Uncharacterised protein family (UPF0497)] |
0.379 |
0.051 |
| 124 |
246498_at |
AT5G16230
|
[Plant stearoyl-acyl-carrier-protein desaturase family protein] |
0.378 |
-0.225 |
| 125 |
264505_at |
AT1G09380
|
UMAMIT25, Usually multiple acids move in and out Transporters 25 |
0.378 |
0.023 |
| 126 |
262452_at |
AT1G11210
|
unknown |
0.377 |
-0.199 |
| 127 |
258957_at |
AT3G01420
|
ALPHA-DOX1, alpha-dioxygenase 1, DIOX1, DOX1, PADOX-1, plant alpha dioxygenase 1 |
0.376 |
0.019 |
| 128 |
246926_at |
AT5G25240
|
unknown |
0.376 |
0.175 |
| 129 |
251774_at |
AT3G55840
|
[Hs1pro-1 protein] |
0.376 |
-0.260 |
| 130 |
251019_at |
AT5G02420
|
unknown |
0.374 |
0.146 |
| 131 |
262516_at |
AT1G17190
|
ATGSTU26, glutathione S-transferase tau 26, GSTU26, glutathione S-transferase tau 26 |
0.372 |
-0.046 |
| 132 |
247359_at |
AT5G63560
|
FACT, FATTY ALCOHOL:CAFFEOYL-CoA CAFFEOYL TRANSFERASE |
0.371 |
0.067 |
| 133 |
255742_at |
AT1G25560
|
AtTEM1, EDF1, ETHYLENE RESPONSE DNA BINDING FACTOR 1, TEM1, TEMPRANILLO 1 |
0.369 |
-0.055 |
| 134 |
265847_at |
AT2G35750
|
unknown |
0.367 |
0.096 |
| 135 |
254001_at |
AT4G26260
|
MIOX4, myo-inositol oxygenase 4 |
0.365 |
0.073 |
| 136 |
258209_at |
AT3G14060
|
unknown |
0.365 |
0.147 |
| 137 |
252421_at |
AT3G47540
|
[Chitinase family protein] |
0.364 |
0.074 |
| 138 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.363 |
0.024 |
| 139 |
255345_at |
AT4G04460
|
[Saposin-like aspartyl protease family protein] |
0.363 |
0.115 |
| 140 |
250857_at |
AT5G04790
|
unknown |
0.361 |
0.048 |
| 141 |
257540_at |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
0.360 |
0.232 |
| 142 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
0.358 |
0.121 |
| 143 |
251183_at |
AT3G62630
|
unknown |
0.358 |
0.044 |
| 144 |
252234_at |
AT3G49780
|
ATPSK3 (FORMER SYMBOL), ATPSK4, phytosulfokine 4 precursor, PSK4, phytosulfokine 4 precursor |
0.353 |
0.061 |
| 145 |
252057_at |
AT3G52480
|
unknown |
0.353 |
0.171 |
| 146 |
252145_at |
AT3G51200
|
SAUR18, SMALL AUXIN UPREGULATED RNA 18 |
0.353 |
0.023 |
| 147 |
259445_at |
AT1G02400
|
ATGA2OX4, Arabidopsis thaliana gibberellin 2-oxidase 4, ATGA2OX6, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 6, DTA1, DOWNSTREAM TARGET OF AGL15 1, GA2OX6, gibberellin 2-oxidase 6 |
0.351 |
-0.189 |
| 148 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
0.351 |
0.003 |
| 149 |
257514_at |
AT1G43940
|
unknown |
0.350 |
0.021 |
| 150 |
255690_at |
AT4G00360
|
ATT1, ABERRANT INDUCTION OF TYPE THREE 1, CYP86A2, cytochrome P450, family 86, subfamily A, polypeptide 2 |
0.348 |
0.054 |
| 151 |
256964_at |
AT3G13520
|
AGP12, arabinogalactan protein 12, ATAGP12 |
0.348 |
0.032 |
| 152 |
245310_at |
AT4G13950
|
RALFL31, ralf-like 31 |
0.348 |
0.042 |
| 153 |
253790_at |
AT4G28660
|
PSB28, photosystem II reaction center PSB28 protein |
0.342 |
0.022 |
| 154 |
249548_at |
AT5G38170
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.342 |
0.021 |
| 155 |
247024_at |
AT5G66985
|
unknown |
0.341 |
0.159 |
| 156 |
251339_at |
AT3G60780
|
unknown |
0.340 |
0.047 |
| 157 |
250563_at |
AT5G08050
|
unknown |
0.340 |
0.017 |
| 158 |
256705_at |
AT3G30290
|
CYP702A8, cytochrome P450, family 702, subfamily A, polypeptide 8 |
0.339 |
0.009 |
| 159 |
248481_at |
AT5G50930
|
[Histone superfamily protein] |
0.339 |
-0.032 |
| 160 |
247111_at |
AT5G65880
|
unknown |
0.338 |
0.039 |
| 161 |
255527_at |
AT4G02360
|
unknown |
0.338 |
0.074 |
| 162 |
259854_at |
AT1G72200
|
[RING/U-box superfamily protein] |
0.337 |
0.175 |
| 163 |
254783_at |
AT4G12830
|
[alpha/beta-Hydrolases superfamily protein] |
0.337 |
-0.205 |
| 164 |
263043_at |
AT1G23350
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.337 |
0.044 |
| 165 |
253024_at |
AT4G38080
|
[hydroxyproline-rich glycoprotein family protein] |
0.334 |
0.062 |
| 166 |
248213_at |
AT5G53660
|
AtGRF7, growth-regulating factor 7, GRF7, growth-regulating factor 7 |
0.333 |
0.056 |
| 167 |
261075_at |
AT1G07280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.331 |
-0.024 |
| 168 |
248406_at |
AT5G51490
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.331 |
0.081 |
| 169 |
251590_at |
AT3G57690
|
AGP23, arabinogalactan protein 23, ATAGP23, ARABINOGALACTAN-PROTEIN 23 |
0.329 |
0.021 |
| 170 |
250281_at |
AT5G13240
|
[transcription regulators] |
0.329 |
0.147 |
| 171 |
261059_at |
AT1G01250
|
[Integrase-type DNA-binding superfamily protein] |
0.328 |
0.080 |
| 172 |
257242_at |
AT3G24220
|
ATNCED6, NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 6, NCED6, nine-cis-epoxycarotenoid dioxygenase 6 |
0.328 |
0.011 |
| 173 |
252214_at |
AT3G50260
|
ATERF#011, CEJ1, cooperatively regulated by ethylene and jasmonate 1, DEAR1, DREB AND EAR MOTIF PROTEIN 1 |
0.327 |
-0.032 |
| 174 |
264680_at |
AT1G65510
|
unknown |
0.325 |
0.097 |
| 175 |
260637_at |
AT1G62380
|
ACO2, ACC oxidase 2, ATACO2 |
0.325 |
-0.026 |
| 176 |
265472_at |
AT2G15580
|
[RING/U-box superfamily protein] |
0.324 |
0.030 |
| 177 |
249875_at |
AT5G23120
|
HCF136, HIGH CHLOROPHYLL FLUORESCENCE 136 |
0.324 |
-0.017 |
| 178 |
262148_at |
AT1G52560
|
[HSP20-like chaperones superfamily protein] |
0.320 |
0.063 |
| 179 |
249894_at |
AT5G22580
|
[Stress responsive A/B Barrel Domain] |
0.319 |
0.138 |
| 180 |
264467_at |
AT1G10140
|
[Uncharacterised conserved protein UCP031279] |
0.319 |
-0.011 |
| 181 |
260561_at |
AT2G43580
|
[Chitinase family protein] |
0.319 |
0.095 |
| 182 |
257421_at |
AT1G12030
|
unknown |
0.318 |
0.086 |
| 183 |
265720_at |
AT2G40110
|
[Yippee family putative zinc-binding protein] |
0.318 |
0.083 |
| 184 |
245305_at |
AT4G17215
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.316 |
0.113 |
| 185 |
260444_at |
AT1G68300
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.316 |
0.060 |
| 186 |
251060_at |
AT5G01820
|
ATCIPK14, ATSR1, serine/threonine protein kinase 1, CIPK14, CBL-INTERACTING PROTEIN KINASE 14, PKS24, SOS2-like protein kinase 24, SnRK3.15, SNF1-RELATED PROTEIN KINASE 3.15, SR1, serine/threonine protein kinase 1 |
0.314 |
0.017 |
| 187 |
258432_at |
AT3G16570
|
ATRALF23, ARABIDOPSIS RAPID ALKALINIZATION FACTOR 23, RALF23, rapid alkalinization factor 23 |
0.314 |
0.070 |
| 188 |
249009_at |
AT5G44610
|
MAP18, microtubule-associated protein 18, PCAP2, PLASMA MEMBRANE ASSOCIATED CA2+-BINDING PROTEIN-2 |
0.313 |
0.090 |
| 189 |
261007_at |
AT1G26400
|
[FAD-binding Berberine family protein] |
0.313 |
0.042 |
| 190 |
262098_at |
AT1G56170
|
ATHAP5B, HAP5B, NF-YC2, nuclear factor Y, subunit C2 |
0.311 |
-0.023 |
| 191 |
249988_at |
AT5G18310
|
unknown |
0.311 |
0.004 |
| 192 |
245998_at |
AT5G20830
|
ASUS1, atsus1, SUS1, sucrose synthase 1 |
0.310 |
0.064 |
| 193 |
246784_at |
AT5G27430
|
[Signal peptidase subunit] |
0.309 |
0.007 |
| 194 |
256215_at |
AT1G50900
|
GDC1, Grana Deficient Chloroplast 1, LTD, LHCP translocation defect |
0.309 |
0.039 |
| 195 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
0.308 |
-0.019 |
| 196 |
265022_at |
AT1G24520
|
BCP1, homolog of Brassica campestris pollen protein 1 |
0.307 |
0.098 |
| 197 |
252037_at |
AT3G51920
|
ATCML9, CAM9, calmodulin 9, CML9, CALMODULIN LIKE PROTEIN 9 |
0.307 |
-0.022 |
| 198 |
256514_at |
AT1G66130
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.306 |
0.002 |
| 199 |
253049_at |
AT4G37300
|
MEE59, maternal effect embryo arrest 59 |
0.303 |
0.046 |
| 200 |
262760_at |
AT1G10770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.303 |
0.053 |
| 201 |
247214_at |
AT5G64850
|
unknown |
0.303 |
-0.081 |
| 202 |
251228_at |
AT3G62710
|
[Glycosyl hydrolase family protein] |
0.303 |
0.037 |
| 203 |
260888_at |
AT1G29140
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.303 |
-0.013 |
| 204 |
247908_at |
AT5G57440
|
GPP2, GLYCEROL-3-PHOSPHATASE 2, GS1 |
0.303 |
0.037 |
| 205 |
260555_at |
AT2G41780
|
unknown |
0.303 |
0.082 |
| 206 |
257584_at |
AT1G54955
|
unknown |
0.302 |
0.083 |
| 207 |
245803_at |
AT1G47128
|
RD21, responsive to dehydration 21, RD21A, responsive to dehydration 21A |
0.301 |
0.006 |
| 208 |
262870_at |
AT1G64710
|
[GroES-like zinc-binding dehydrogenase family protein] |
0.301 |
0.047 |
| 209 |
254187_at |
AT4G23890
|
CRR31, CHLORORESPIRATORY REDUCTION 31, NdhS, NADH dehydrogenase-like complex S |
0.300 |
-0.011 |
| 210 |
257775_at |
AT3G29260
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.300 |
0.018 |
| 211 |
254059_at |
AT4G25200
|
ATHSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6, HSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6 |
0.300 |
0.121 |
| 212 |
248419_at |
AT5G51550
|
EXL3, EXORDIUM like 3 |
0.300 |
0.064 |
| 213 |
248167_at |
AT5G54530
|
unknown |
0.299 |
0.073 |
| 214 |
248214_at |
AT5G53670
|
unknown |
0.299 |
0.029 |
| 215 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
0.297 |
0.048 |
| 216 |
264262_at |
AT1G09200
|
H3.1, histone 3.1 |
0.297 |
-0.003 |
| 217 |
247295_at |
AT5G64180
|
unknown |
0.296 |
0.113 |
| 218 |
256518_at |
AT1G66080
|
unknown |
0.296 |
0.123 |
| 219 |
264098_at |
AT1G79260
|
unknown |
0.296 |
0.012 |
| 220 |
263369_at |
AT2G20480
|
unknown |
0.295 |
0.115 |
| 221 |
263689_at |
AT1G26820
|
RNS3, ribonuclease 3 |
0.295 |
0.115 |
| 222 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
0.295 |
-0.553 |
| 223 |
260887_at |
AT1G29160
|
[Dof-type zinc finger DNA-binding family protein] |
0.295 |
0.220 |
| 224 |
251545_at |
AT3G58810
|
ATMTP3, ARABIDOPSIS METAL TOLERANCE PROTEIN 3, ATMTPA2, MTP3, METAL TOLERANCE PROTEIN 3, MTPA2, metal tolerance protein A2 |
0.294 |
0.134 |
| 225 |
260871_at |
AT1G29040
|
unknown |
0.293 |
0.095 |
| 226 |
245629_at |
AT1G56580
|
SVB, SMALLER WITH VARIABLE BRANCHES |
0.292 |
-0.035 |
| 227 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
0.291 |
0.207 |
| 228 |
253660_at |
AT4G30140
|
CDEF1, CUTICLE DESTRUCTING FACTOR 1 |
0.290 |
-0.134 |
| 229 |
250058_at |
AT5G17870
|
PSRP6, plastid-specific 50S ribosomal protein 6 |
0.290 |
0.044 |
| 230 |
261441_at |
AT1G28470
|
ANAC010, NAC domain containing protein 10, NAC010, NAC domain containing protein 10, SND3, SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN 3 |
0.289 |
0.042 |
| 231 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
0.289 |
-0.037 |
| 232 |
251323_at |
AT3G61580
|
AtSLD1, SLD1, sphingoid LCB desaturase 1 |
0.288 |
-0.003 |
| 233 |
251961_at |
AT3G53620
|
AtPPa4, pyrophosphorylase 4, PPa4, pyrophosphorylase 4 |
0.288 |
0.177 |
| 234 |
253606_at |
AT4G30530
|
GGP1, gamma-glutamyl peptidase 1 |
0.287 |
-0.004 |
| 235 |
249576_at |
AT5G37690
|
[SGNH hydrolase-type esterase superfamily protein] |
0.287 |
0.023 |
| 236 |
252502_at |
AT3G46900
|
COPT2, copper transporter 2 |
0.287 |
0.054 |
| 237 |
266131_at |
AT2G45160
|
ATHAM1, ARABIDOPSIS THALIANA HAIRY MERISTEM 1, HAM1, HAIRY MERISTEM 1, LOM1, LOST MERISTEMS 1 |
0.287 |
0.012 |
| 238 |
264436_at |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
0.286 |
0.002 |
| 239 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.286 |
0.041 |
| 240 |
245367_at |
AT4G16265
|
NRPB9B, NRPD9B, NRPE9B |
0.286 |
0.032 |
| 241 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
0.285 |
0.283 |
| 242 |
251513_at |
AT3G59220
|
ATPIRIN1, PRN, pirin, PRN1 |
0.285 |
-0.005 |
| 243 |
245243_at |
AT1G44414
|
unknown |
0.285 |
0.080 |
| 244 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.285 |
0.014 |
| 245 |
262719_at |
AT1G43590
|
unknown |
0.284 |
-0.001 |
| 246 |
252036_at |
AT3G52070
|
unknown |
0.284 |
0.106 |
| 247 |
254201_at |
AT4G24130
|
unknown |
0.284 |
0.020 |
| 248 |
248822_at |
AT5G47000
|
[Peroxidase superfamily protein] |
0.282 |
0.041 |
| 249 |
260438_at |
AT1G68290
|
ENDO2, endonuclease 2 |
0.282 |
-0.105 |
| 250 |
248504_at |
AT5G50480
|
NF-YC6, nuclear factor Y, subunit C6 |
0.282 |
0.019 |
| 251 |
260391_at |
AT1G74020
|
SS2, strictosidine synthase 2 |
0.281 |
0.068 |
| 252 |
250537_at |
AT5G08565
|
[Transcription initiation Spt4-like protein] |
0.281 |
0.155 |
| 253 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
0.281 |
0.061 |
| 254 |
247964_at |
AT5G56600
|
PFN3, PROFILIN 3, PRF3, profilin 3 |
0.281 |
0.072 |
| 255 |
252133_at |
AT3G50900
|
unknown |
0.280 |
-0.046 |
| 256 |
252560_at |
AT3G46030
|
HTB11 |
0.279 |
-0.060 |
| 257 |
253391_at |
AT4G32590
|
[2Fe-2S ferredoxin-like superfamily protein] |
0.279 |
0.115 |
| 258 |
259179_at |
AT3G01660
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.278 |
-0.055 |
| 259 |
265032_at |
AT1G61580
|
ARP2, ARABIDOPSIS RIBOSOMAL PROTEIN 2, RPL3B, R-protein L3 B |
0.278 |
-0.022 |
| 260 |
267178_at |
AT2G37750
|
unknown |
0.278 |
0.102 |
| 261 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
0.277 |
0.150 |
| 262 |
261410_at |
AT1G07610
|
MT1C, metallothionein 1C |
0.277 |
0.018 |
| 263 |
251369_at |
AT3G60480
|
unknown |
0.276 |
0.113 |
| 264 |
261109_at |
AT1G75450
|
ATCKX5, ARABIDOPSIS THALIANA CYTOKININ OXIDASE 5, ATCKX6, CYTOKININ OXIDASE 6, CKX5, cytokinin oxidase 5 |
0.276 |
0.086 |
| 265 |
264188_at |
AT1G54690
|
G-H2AX, GAMMA H2AX, GAMMA-H2AX, gamma histone variant H2AX, H2AXB, HTA3, histone H2A 3 |
0.276 |
-0.021 |
| 266 |
257483_at |
AT1G49620
|
AtKRP7, ICK5, ICN6, KRP7, KIP-RELATED PROTEIN 7 |
0.275 |
-0.025 |
| 267 |
247646_at |
AT5G59990
|
[CCT motif family protein] |
0.275 |
0.058 |
| 268 |
258143_at |
AT3G18170
|
[Glycosyltransferase family 61 protein] |
0.275 |
0.028 |
| 269 |
245238_at |
AT4G25570
|
ACYB-2 |
0.275 |
0.046 |
| 270 |
264887_at |
AT1G23120
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.275 |
0.062 |
| 271 |
264209_at |
AT1G22740
|
ATRABG3B, RAB7, RAB75, RABG3B, RAB GTPase homolog G3B |
0.274 |
0.023 |
| 272 |
260850_at |
AT1G21870
|
GONST5, golgi nucleotide sugar transporter 5 |
0.274 |
0.055 |
| 273 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
0.273 |
-0.173 |
| 274 |
252024_at |
AT3G52880
|
ATMDAR1, monodehydroascorbate reductase 1, MDAR1, monodehydroascorbate reductase 1 |
0.273 |
-0.016 |
| 275 |
253181_at |
AT4G35180
|
LHT7, LYS/HIS transporter 7 |
0.272 |
-0.034 |
| 276 |
255904_at |
AT1G17860
|
[Kunitz family trypsin and protease inhibitor protein] |
0.272 |
-0.031 |
| 277 |
256053_at |
AT1G07260
|
UGT71C3, UDP-glucosyl transferase 71C3 |
0.271 |
-0.002 |
| 278 |
261269_at |
AT1G26690
|
[emp24/gp25L/p24 family/GOLD family protein] |
0.271 |
0.075 |
| 279 |
250110_at |
AT5G15350
|
AtENODL17, ENODL17, early nodulin-like protein 17 |
0.270 |
0.016 |
| 280 |
253859_at |
AT4G27657
|
unknown |
0.269 |
-0.472 |
| 281 |
252761_at |
AT3G42770
|
[F-box/RNI-like/FBD-like domains-containing protein] |
0.269 |
0.058 |
| 282 |
245096_at |
AT2G40880
|
ATCYSA, cystatin A, CYSA, cystatin A, FL3-27 |
0.269 |
-0.021 |
| 283 |
252005_at |
AT3G52810
|
ATPAP21, PURPLE ACID PHOSPHATASE 21, PAP21, purple acid phosphatase 21 |
0.269 |
0.067 |
| 284 |
247774_at |
AT5G58660
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.268 |
0.010 |
| 285 |
267591_at |
AT2G39705
|
DVL11, DEVIL 11, RTFL8, ROTUNDIFOLIA like 8 |
0.268 |
0.109 |
| 286 |
251858_at |
AT3G54820
|
PIP2;5, plasma membrane intrinsic protein 2;5, PIP2D, PLASMA MEMBRANE INTRINSIC PROTEIN 2D |
0.267 |
0.042 |
| 287 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
0.267 |
-0.199 |
| 288 |
264708_at |
AT1G09740
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.265 |
0.068 |
| 289 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.265 |
-0.179 |
| 290 |
261791_at |
AT1G16170
|
unknown |
0.264 |
0.061 |
| 291 |
248200_at |
AT5G54160
|
ATOMT1, O-methyltransferase 1, COMT1, caffeate O-methyltransferase 1, OMT1, O-methyltransferase 1 |
0.263 |
-0.027 |
| 292 |
255691_at |
AT4G00370
|
ANTR2, PHT4;4, anion transporter 2 |
0.263 |
-0.033 |
| 293 |
254559_at |
AT4G19200
|
[proline-rich family protein] |
0.263 |
0.024 |
| 294 |
249547_at |
AT5G38160
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.263 |
0.032 |
| 295 |
257635_at |
AT3G26280
|
CYP71B4, cytochrome P450, family 71, subfamily B, polypeptide 4 |
0.262 |
0.257 |
| 296 |
265983_at |
AT2G18550
|
ATHB21, homeobox protein 21, HB-2, homeobox-2, HB21, homeobox protein 21 |
0.262 |
0.026 |
| 297 |
262838_at |
AT1G14960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.262 |
-0.010 |
| 298 |
250228_at |
AT5G13840
|
CCS52B, cell cycle switch protein 52 B, FZR3, FIZZY-related 3 |
0.262 |
-0.070 |
| 299 |
266617_at |
AT2G29670
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.261 |
0.013 |
| 300 |
252946_at |
AT4G39235
|
unknown |
0.261 |
0.095 |
| 301 |
247714_at |
AT5G59340
|
WOX2, WUSCHEL related homeobox 2 |
-0.773 |
0.089 |
| 302 |
244936_at |
ATCG01100
|
NDHA |
-0.699 |
-0.075 |
| 303 |
244935_at |
ATCG01090
|
NDHI |
-0.690 |
-0.032 |
| 304 |
265475_at |
AT2G15620
|
ATHNIR, ARABIDOPSIS THALIANA NITRITE REDUCTASE, NIR, NITRITE REDUCTASE, NIR1, nitrite reductase 1 |
-0.648 |
-0.028 |
| 305 |
264101_at |
AT1G79000
|
ATHAC1, ARABIDOPSIS HISTONE ACETYLTRANSFERASE OF THE CBP FAMILY 1, ATHPCAT2, ARABIDOPSIS THALIANA P300/CBP ACETYLTRANSFERASE-RELATED PROTEIN 2, HAC1, histone acetyltransferase of the CBP family 1, PCAT2, P300/CBP ACETYLTRANSFERASE-RELATED PROTEIN 2 |
-0.646 |
-0.084 |
| 306 |
258223_at |
AT3G15840
|
PIFI, post-illumination chlorophyll fluorescence increase |
-0.646 |
0.034 |
| 307 |
252092_at |
AT3G51420
|
ATSSL4, STRICTOSIDINE SYNTHASE-LIKE 4, SSL4, strictosidine synthase-like 4 |
-0.624 |
0.068 |
| 308 |
263192_at |
AT1G36160
|
ACC1, acetyl-CoA carboxylase 1, AT-ACC1, EMB22, EMBRYO DEFECTIVE 22, GK, GURKE, GSD1, GLOSSYHEAD 1, PAS3, PASTICCINO 3, SFR3, SENSITIVE TO FREEZING 3 |
-0.598 |
-0.058 |
| 309 |
246095_at |
AT5G19310
|
CHR23, chromatin remodeling 23 |
-0.576 |
-0.394 |
| 310 |
248673_at |
AT5G48780
|
[disease resistance protein (TIR-NBS class)] |
-0.569 |
-0.062 |
| 311 |
253975_at |
AT4G26600
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.561 |
-0.517 |
| 312 |
267160_at |
AT2G37670
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.560 |
0.017 |
| 313 |
256675_at |
AT3G52170
|
[DNA binding] |
-0.553 |
-0.291 |
| 314 |
257273_at |
AT3G28030
|
UVH3, ULTRAVIOLET HYPERSENSITIVE 3, UVR1, UV REPAIR DEFECTIVE 1 |
-0.552 |
-0.203 |
| 315 |
247921_at |
AT5G57660
|
ATCOL5, BBX6, B-box domain protein 6, COL5, CONSTANS-like 5 |
-0.548 |
0.090 |
| 316 |
260459_at |
AT1G68240
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.547 |
0.050 |
| 317 |
248939_at |
AT5G45790
|
[Ubiquitin carboxyl-terminal hydrolase family protein] |
-0.541 |
-0.096 |
| 318 |
254944_at |
AT4G10930
|
unknown |
-0.539 |
-0.224 |
| 319 |
247867_at |
AT5G57630
|
CIPK21, CBL-interacting protein kinase 21, SnRK3.4, SNF1-RELATED PROTEIN KINASE 3.4 |
-0.536 |
-0.200 |
| 320 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
-0.535 |
-0.194 |
| 321 |
253458_at |
AT4G32070
|
Phox4, Phox4 |
-0.534 |
-0.056 |
| 322 |
258157_at |
AT3G18100
|
AtMYB4R1, myb domain protein 4R1, MYB4R1, myb domain protein 4r1 |
-0.530 |
-0.223 |
| 323 |
249409_at |
AT5G40340
|
[Tudor/PWWP/MBT superfamily protein] |
-0.520 |
-0.194 |
| 324 |
264390_at |
AT1G11950
|
[Transcription factor jumonji (jmjC) domain-containing protein] |
-0.517 |
-0.177 |
| 325 |
260627_at |
AT1G62310
|
[transcription factor jumonji (jmjC) domain-containing protein] |
-0.510 |
-0.146 |
| 326 |
259432_at |
AT1G01520
|
ASG4, ALTERED SEED GERMINATION 4 |
-0.508 |
-0.019 |
| 327 |
250601_at |
AT5G07810
|
[SNF2 domain-containing protein / helicase domain-containing protein / HNH endonuclease domain-containing protein] |
-0.508 |
-0.205 |
| 328 |
246055_at |
AT5G08380
|
AGAL1, alpha-galactosidase 1, AtAGAL1, alpha-galactosidase 1 |
-0.502 |
0.156 |
| 329 |
264471_at |
AT1G67120
|
AtMDN1, MDN1, MIDASIN 1 |
-0.497 |
-0.248 |
| 330 |
266361_at |
AT2G32450
|
[Calcium-binding tetratricopeptide family protein] |
-0.494 |
-0.027 |
| 331 |
258833_at |
AT3G07274
|
|
-0.494 |
-0.186 |
| 332 |
249020_at |
AT5G44800
|
CHR4, chromatin remodeling 4, PKR1, PICKLE RELATED 1 |
-0.490 |
-0.096 |
| 333 |
249426_at |
AT5G39840
|
[ATP-dependent RNA helicase, mitochondrial, putative] |
-0.487 |
-0.099 |
| 334 |
261333_at |
AT1G44910
|
ATPRP40A, pre-mRNA-processing protein 40A, PRP40A, pre-mRNA-processing protein 40A |
-0.483 |
-0.392 |
| 335 |
258333_at |
AT3G16000
|
MFP1, MAR binding filament-like protein 1 |
-0.482 |
-0.348 |
| 336 |
263448_at |
AT2G31660
|
EMA1, enhanced miRNA activity 1, SAD2, SUPER SENSITIVE TO ABA AND DROUGHT2, URM9, UNARMED 9 |
-0.479 |
-0.021 |
| 337 |
245585_at |
AT4G14970
|
unknown |
-0.478 |
-0.070 |
| 338 |
262614_at |
AT1G13980
|
EMB30, EMBRYO DEFECTIVE 30, GN, GNOM, VAN7, VASCULAR NETWORK 7 |
-0.475 |
-0.060 |
| 339 |
261806_at |
AT1G30510
|
ATRFNR2, root FNR 2, RFNR2, root FNR 2 |
-0.474 |
-0.051 |
| 340 |
259681_at |
AT1G77760
|
GNR1, NIA1, nitrate reductase 1, NR1, NITRATE REDUCTASE 1 |
-0.473 |
-0.025 |
| 341 |
259794_at |
AT1G64330
|
[myosin heavy chain-related] |
-0.473 |
-0.331 |
| 342 |
266866_at |
AT2G29940
|
ABCG31, ATP-binding cassette G31, ATPDR3, PLEIOTROPIC DRUG RESISTANCE 3, PDR3, pleiotropic drug resistance 3 |
-0.473 |
-0.022 |
| 343 |
249843_at |
AT5G23570
|
ATSGS3, SUPPRESSOR OF GENE SILENCING 3, SGS3, SUPPRESSOR OF GENE SILENCING 3 |
-0.473 |
-0.112 |
| 344 |
247142_at |
AT5G65570
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.472 |
-0.066 |
| 345 |
250087_at |
AT5G17270
|
[Protein prenylyltransferase superfamily protein] |
-0.469 |
-0.253 |
| 346 |
261494_at |
AT1G28420
|
HB-1, homeobox-1, RLT1, RINGLET 1 |
-0.469 |
-0.151 |
| 347 |
265678_at |
AT2G31970
|
ATRAD50, RAD50 |
-0.469 |
-0.175 |
| 348 |
247032_at |
AT5G67240
|
SDN3, small RNA degrading nuclease 3 |
-0.459 |
-0.400 |
| 349 |
257026_at |
AT3G19200
|
unknown |
-0.457 |
-0.012 |
| 350 |
261923_at |
AT1G22380
|
AtUGT85A3, UDP-glucosyl transferase 85A3, UGT85A3, UDP-glucosyl transferase 85A3 |
-0.456 |
0.071 |
| 351 |
247191_at |
AT5G65310
|
ATHB-5, ATHB5, homeobox protein 5, HB5, homeobox protein 5 |
-0.456 |
-0.030 |
| 352 |
252411_at |
AT3G47430
|
PEX11B, peroxin 11B |
-0.455 |
-0.012 |
| 353 |
262323_at |
AT1G64190
|
[6-phosphogluconate dehydrogenase family protein] |
-0.455 |
0.016 |
| 354 |
246383_at |
AT1G77360
|
APPR6, Arabidopsis pentatricopeptide repeat 6 |
-0.454 |
-0.047 |
| 355 |
260177_at |
AT1G70650
|
[Ran BP2/NZF zinc finger-like superfamily protein] |
-0.454 |
-0.158 |
| 356 |
246120_at |
AT5G20360
|
Phox3, Phox3 |
-0.450 |
-0.190 |
| 357 |
262988_at |
AT1G23310
|
AOAT1, ALANINE-2-OXOGLUTARATE AMINOTRANSFERASE 1, GGAT1, GLUTAMATE:GLYOXYLATE AMINOTRANSFERASE 1, GGT1, glutamate:glyoxylate aminotransferase |
-0.439 |
-0.028 |
| 358 |
263605_at |
AT2G16485
|
NERD, Needed for RDR2-independent DNA methylation |
-0.438 |
-0.181 |
| 359 |
263349_at |
AT2G13370
|
CHR5, chromatin remodeling 5 |
-0.438 |
-0.098 |
| 360 |
246558_at |
AT5G15540
|
ATSCC2, ARABIDOPSIS THALIANA SISTER-CHROMATID COHESION 2, EMB2773, EMBRYO DEFECTIVE 2773, SCC2, SISTER-CHROMATID COHESION 2 |
-0.438 |
-0.156 |
| 361 |
260031_at |
AT1G68790
|
CRWN3, CROWDED NUCLEI 3, LINC3, LITTLE NUCLEI3 |
-0.437 |
-0.348 |
| 362 |
266009_at |
AT2G37420
|
[ATP binding microtubule motor family protein] |
-0.437 |
-0.157 |
| 363 |
260717_at |
AT1G48120
|
[hydrolases] |
-0.430 |
-0.086 |
| 364 |
261857_at |
AT1G50620
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-0.426 |
-0.161 |
| 365 |
246731_at |
AT5G27630
|
ACBP5, acyl-CoA binding protein 5, AtACBP5 |
-0.421 |
-0.014 |
| 366 |
248307_at |
AT5G52850
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.419 |
-0.172 |
| 367 |
266002_at |
AT2G37310
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.418 |
-0.023 |
| 368 |
248994_at |
AT5G45250
|
RPS4, RESISTANT TO P. SYRINGAE 4 |
-0.417 |
-0.227 |
| 369 |
257982_at |
AT3G20780
|
ATTOP6B, topoisomerase 6 subunit B, BIN3, BRASSINOSTEROID INSENSITIVE 3, HYP6, ELONGATED HYPOCOTYL 6, RHL3, ROOT HAIRLESS 3, TOP6B, topoisomerase 6 subunit B |
-0.417 |
-0.250 |
| 370 |
256332_at |
AT1G76890
|
AT-GT2, GT2 |
-0.415 |
-0.304 |
| 371 |
259899_at |
AT1G71210
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.414 |
-0.142 |
| 372 |
245709_at |
AT5G04320
|
[Shugoshin C terminus] |
-0.414 |
-0.391 |
| 373 |
266050_at |
AT2G40770
|
[zinc ion binding] |
-0.412 |
-0.163 |
| 374 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
-0.411 |
0.022 |
| 375 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.409 |
-0.059 |
| 376 |
259872_at |
AT1G76810
|
[eukaryotic translation initiation factor 2 (eIF-2) family protein] |
-0.408 |
-0.175 |
| 377 |
248874_at |
AT5G46460
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.408 |
-0.052 |
| 378 |
253532_at |
AT4G31570
|
unknown |
-0.407 |
-0.139 |
| 379 |
258075_at |
AT3G25900
|
ATHMT-1, HMT-1 |
-0.407 |
0.052 |
| 380 |
267508_at |
AT2G45700
|
[sterile alpha motif (SAM) domain-containing protein] |
-0.406 |
0.022 |
| 381 |
249115_at |
AT5G43810
|
AGO10, ARGONAUTE 10, PNH, PINHEAD, ZLL, ZWILLE |
-0.406 |
-0.034 |
| 382 |
266778_at |
AT2G29090
|
CYP707A2, cytochrome P450, family 707, subfamily A, polypeptide 2 |
-0.405 |
-0.269 |
| 383 |
248465_at |
AT5G51200
|
EMB3142, EMBRYO DEFECTIVE 3142 |
-0.405 |
-0.043 |
| 384 |
265326_at |
AT2G18220
|
[Noc2p family] |
-0.403 |
-0.391 |
| 385 |
248315_at |
AT5G52630
|
MEF1, mitochondrial RNA editing factor 1 |
-0.402 |
-0.099 |
| 386 |
258471_at |
AT3G06030
|
ANP3, NPK1-related protein kinase 3, AtANP3, MAPKKK12, NP3, NPK1-related protein kinase 3 |
-0.401 |
-0.052 |
| 387 |
256659_at |
AT3G12020
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.399 |
-0.241 |
| 388 |
246261_at |
AT1G31810
|
AFH14, Formin Homology 14 |
-0.399 |
-0.110 |
| 389 |
263909_at |
AT2G36490
|
AtROS1, DML1, demeter-like 1, ROS1, REPRESSOR OF SILENCING1 |
-0.399 |
-0.072 |
| 390 |
262001_at |
AT1G33790
|
[jacalin lectin family protein] |
-0.398 |
-0.039 |
| 391 |
261086_at |
AT1G17460
|
TRFL3, TRF-like 3 |
-0.397 |
-0.183 |
| 392 |
247085_at |
AT5G66310
|
[ATP binding microtubule motor family protein] |
-0.396 |
-0.189 |
| 393 |
262362_at |
AT1G72840
|
[Disease resistance protein (TIR-NBS-LRR class)] |
-0.396 |
0.027 |
| 394 |
262766_at |
AT1G13160
|
[ARM repeat superfamily protein] |
-0.395 |
-0.329 |
| 395 |
264790_at |
AT2G17820
|
AHK1, ATHK1, histidine kinase 1, HK1, histidine kinase 1 |
-0.395 |
-0.123 |
| 396 |
260282_at |
AT1G80410
|
EMB2753, EMBRYO DEFECTIVE 2753, OMA, OMISHA |
-0.395 |
-0.091 |
| 397 |
257007_at |
AT3G14205
|
[Phosphoinositide phosphatase family protein] |
-0.394 |
-0.237 |
| 398 |
249292_at |
AT5G41150
|
ATRAD1, RAD1, UVH1, ULTRAVIOLET HYPERSENSITIVE 1 |
-0.393 |
-0.024 |
| 399 |
260406_at |
AT1G69920
|
ATGSTU12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 12, GSTU12, glutathione S-transferase TAU 12 |
-0.393 |
-0.068 |
| 400 |
264948_at |
AT1G77030
|
[hydrolases, acting on acid anhydrides, in phosphorus-containing anhydrides] |
-0.393 |
-0.205 |
| 401 |
256661_at |
AT3G11964
|
RRP5, Ribosomal RNA Processing 5 |
-0.392 |
-0.188 |
| 402 |
253907_at |
AT4G27250
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.390 |
0.054 |
| 403 |
254238_at |
AT4G23540
|
[ARM repeat superfamily protein] |
-0.389 |
-0.328 |
| 404 |
250788_at |
AT5G05570
|
[transducin family protein / WD-40 repeat family protein] |
-0.386 |
-0.059 |
| 405 |
261447_at |
AT1G21160
|
[eukaryotic translation initiation factor 2 (eIF-2) family protein] |
-0.384 |
-0.115 |
| 406 |
256613_at |
AT3G29290
|
emb2076, embryo defective 2076 |
-0.382 |
-0.121 |
| 407 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
-0.382 |
0.055 |
| 408 |
258559_at |
AT3G06010
|
ATCHR12, CHR12, chromatin remodeling 12 |
-0.381 |
-0.286 |
| 409 |
253133_at |
AT4G35800
|
NRPB1, RNA polymerase II large subunit, RNA_POL_II_LS, RNA POLYMERASE II LARGE SUBUNIT, RNA_POL_II_LSRNA_POL_II_LS, RPB1, RNA POLYMERASE II LARGE SUBUNIT |
-0.380 |
-0.018 |
| 410 |
244983_at |
ATCG00790
|
RPL16, ribosomal protein L16 |
-0.380 |
-0.463 |
| 411 |
253083_at |
AT4G36250
|
ALDH3F1, aldehyde dehydrogenase 3F1 |
-0.379 |
-0.072 |
| 412 |
249817_at |
AT5G23820
|
ML3, MD2-related lipid recognition 3 |
-0.379 |
-0.007 |
| 413 |
260641_at |
AT1G53200
|
unknown |
-0.379 |
-0.182 |
| 414 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
-0.375 |
-0.453 |
| 415 |
244996_at |
ATCG00160
|
RPS2, ribosomal protein S2 |
-0.375 |
-0.124 |
| 416 |
260343_at |
AT1G69200
|
FLN2, fructokinase-like 2 |
-0.374 |
-0.259 |
| 417 |
245302_at |
AT4G17695
|
KAN3, KANADI 3 |
-0.374 |
-0.240 |
| 418 |
244986_at |
ATCG00820
|
RPS19, ribosomal protein S19 |
-0.374 |
-0.237 |
| 419 |
261701_at |
AT1G32750
|
GTD1, HAC13, HISTONE ACETYLTRANSFERASE OF THE CBP FAMILY 13, HAF01, HAF1, HISTONE ACETYLTRANSFERASE OF THE TAFII250 FAMILY 1, TAF1, TBP-ASSOCIATED FACTOR 1 |
-0.373 |
-0.300 |
| 420 |
246493_at |
AT5G16180
|
ATCRS1, ARABIDOPSIS ORTHOLOG OF MAIZE CHLOROPLAST SPLICING FACTOR CRS1, CRS1, ortholog of maize chloroplast splicing factor CRS1 |
-0.373 |
-0.175 |
| 421 |
259759_at |
AT1G77550
|
[tubulin-tyrosine ligases] |
-0.372 |
-0.030 |
| 422 |
256575_at |
AT3G14790
|
ATRHM3, ARABIDOPSIS THALIANA RHAMNOSE BIOSYNTHESIS 3, RHM3, rhamnose biosynthesis 3 |
-0.372 |
-0.015 |
| 423 |
253019_at |
AT4G38010
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
-0.372 |
-0.020 |
| 424 |
266659_at |
AT2G25850
|
PAPS2, poly(A) polymerase 2 |
-0.371 |
-0.140 |
| 425 |
260857_at |
AT1G21880
|
LYM1, lysm domain GPI-anchored protein 1 precursor, LYP2, LysM-containing receptor protein 2 |
-0.370 |
-0.017 |
| 426 |
244980_at |
ATCG00760
|
RPL36, ribosomal protein L36 |
-0.370 |
-0.303 |
| 427 |
258048_at |
AT3G16290
|
EMB2083, embryo defective 2083 |
-0.370 |
-0.106 |
| 428 |
250245_at |
AT5G13690
|
CYL1, CYCLOPS 1, NAGLU, N-ACETYL-GLUCOSAMINIDASE |
-0.370 |
-0.108 |
| 429 |
260412_at |
AT1G69830
|
AMY3, alpha-amylase-like 3, ATAMY3, ALPHA-AMYLASE-LIKE 3 |
-0.370 |
-0.078 |
| 430 |
260251_at |
AT1G74250
|
[DNAJ heat shock N-terminal domain-containing protein] |
-0.369 |
-0.389 |
| 431 |
264689_at |
AT1G09900
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
-0.368 |
-0.003 |
| 432 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
-0.367 |
0.132 |
| 433 |
263174_at |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
-0.367 |
0.055 |
| 434 |
252082_at |
AT3G51940
|
unknown |
-0.367 |
0.002 |
| 435 |
262220_at |
AT1G74740
|
ATCPK30, CDPK1A, CALCIUM-DEPENDENT PROTEIN KINASE 1A, CPK30, calcium-dependent protein kinase 30 |
-0.367 |
-0.077 |
| 436 |
246424_at |
AT5G17020
|
ATCRM1, ATXPO1, ARABIDOPSIS THALIANA EXPORTIN 1, HIT2, HEAT-INTOLERANT 2, XPO1, EXPORTIN 1, XPO1A, exportin 1A |
-0.367 |
-0.029 |
| 437 |
246447_at |
AT5G16780
|
DOT2, DEFECTIVELY ORGANIZED TRIBUTARIES 2, MDF, MERISTEM-DEFECTIVE |
-0.363 |
-0.197 |
| 438 |
245109_at |
AT2G41520
|
DJC62, DNA J protein C62, TPR15, tetratricopeptide repeat 15 |
-0.362 |
-0.106 |
| 439 |
262150_at |
AT1G52520
|
FRS6, FAR1-related sequence 6 |
-0.362 |
-0.053 |
| 440 |
267028_at |
AT2G38470
|
ATWRKY33, WRKY DNA-BINDING PROTEIN 33, WRKY33, WRKY DNA-binding protein 33 |
-0.360 |
-0.374 |
| 441 |
252245_at |
AT3G49710
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.359 |
-0.116 |
| 442 |
250374_at |
AT5G11530
|
EMF1, embryonic flower 1 |
-0.359 |
-0.115 |
| 443 |
253144_at |
AT4G35540
|
PTF2, Pollen-expressed Transcription Factor 2 |
-0.359 |
-0.131 |
| 444 |
245509_at |
AT4G15730
|
[CW-type Zinc Finger] |
-0.358 |
0.000 |
| 445 |
261750_at |
AT1G76120
|
[Pseudouridine synthase family protein] |
-0.358 |
-0.142 |
| 446 |
246184_at |
AT5G20950
|
[Glycosyl hydrolase family protein] |
-0.358 |
0.040 |
| 447 |
254724_at |
AT4G13630
|
unknown |
-0.358 |
-0.105 |
| 448 |
254924_at |
AT4G11330
|
ATMPK5, MAP kinase 5, MPK5, MAP kinase 5 |
-0.358 |
-0.026 |
| 449 |
263165_at |
AT1G03060
|
SPI, SPIRRIG |
-0.358 |
-0.063 |
| 450 |
260466_at |
AT1G10900
|
[Phosphatidylinositol-4-phosphate 5-kinase family protein] |
-0.358 |
-0.076 |
| 451 |
261762_at |
AT1G15510
|
ATECB2, ARABIDOPSIS EARLY CHLOROPLAST BIOGENESIS2, ECB2, EARLY CHLOROPLAST BIOGENESIS2, VAC1, VANILLA CREAM 1 |
-0.357 |
-0.142 |
| 452 |
262081_at |
AT1G59540
|
ZCF125 |
-0.356 |
-0.211 |
| 453 |
255522_at |
AT4G02260
|
AT-RSH1, RELA-SPOT HOMOLOG 1, ATRSH1, RELA/SPOT HOMOLOG 1, RSH1, RELA/SPOT homolog 1 |
-0.355 |
-0.122 |
| 454 |
248291_at |
AT5G53020
|
[Ribonuclease P protein subunit P38-related] |
-0.355 |
-0.196 |
| 455 |
262778_at |
AT1G13050
|
unknown |
-0.354 |
0.098 |
| 456 |
261476_at |
AT1G14480
|
[Ankyrin repeat family protein] |
-0.354 |
-0.294 |
| 457 |
249808_at |
AT5G23890
|
[LOCATED IN: mitochondrion, chloroplast thylakoid membrane, chloroplast, plastid, chloroplast envelope] |
-0.354 |
-0.130 |
| 458 |
250164_at |
AT5G15280
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.354 |
-0.078 |
| 459 |
249871_at |
AT5G23110
|
[Zinc finger, C3HC4 type (RING finger) family protein] |
-0.353 |
-0.004 |
| 460 |
245256_at |
AT4G15090
|
FAR1, FAR-RED IMPAIRED RESPONSE 1 |
-0.352 |
-0.120 |
| 461 |
260292_at |
AT1G63680
|
APG13, ALBINO OR PALE-GREEN 13, ATMURE, MURE, PDE316, PIGMENT DEFECTIVE EMBRYO 316 |
-0.352 |
-0.149 |
| 462 |
257516_at |
AT1G69040
|
ACR4, ACT domain repeat 4 |
-0.352 |
0.005 |
| 463 |
245845_at |
AT1G26150
|
AtPERK10, proline-rich extensin-like receptor kinase 10, PERK10, proline-rich extensin-like receptor kinase 10 |
-0.352 |
0.007 |
| 464 |
251593_at |
AT3G57660
|
NRPA1, nuclear RNA polymerase A1 |
-0.352 |
-0.256 |
| 465 |
261761_at |
AT1G15660
|
CENP-C, CENP-C HOMOLOGUE, CENP-C, centromere protein C |
-0.352 |
-0.131 |
| 466 |
256537_at |
AT1G33340
|
[ENTH/ANTH/VHS superfamily protein] |
-0.352 |
0.062 |
| 467 |
249791_at |
AT5G23810
|
AAP7, amino acid permease 7 |
-0.351 |
0.086 |
| 468 |
246162_at |
AT4G36400
|
D2HGDH, D-2-hydroxyglutarate dehydrogenase |
-0.351 |
-0.028 |
| 469 |
253173_at |
AT4G35110
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
-0.350 |
-0.060 |
| 470 |
247603_at |
AT5G60930
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.348 |
-0.232 |
| 471 |
246457_at |
AT5G16750
|
TOZ, TORMOZEMBRYO DEFECTIVE |
-0.347 |
-0.192 |
| 472 |
259303_at |
AT3G05130
|
unknown |
-0.345 |
-0.291 |
| 473 |
255149_at |
AT4G08150
|
BP, BREVIPEDICELLUS, BP1, BREVIPEDICELLUS 1, KNAT1, KNOTTED-like from Arabidopsis thaliana |
-0.345 |
-0.134 |
| 474 |
248933_at |
AT5G46070
|
[Guanylate-binding family protein] |
-0.344 |
-0.305 |
| 475 |
254639_at |
AT4G18750
|
DOT4, DEFECTIVELY ORGANIZED TRIBUTARIES 4 |
-0.343 |
-0.198 |
| 476 |
265467_at |
AT2G37050
|
[Leucine-rich repeat protein kinase family protein] |
-0.342 |
0.036 |
| 477 |
261117_at |
AT1G75310
|
AUL1, auxilin-like 1 |
-0.341 |
-0.183 |
| 478 |
248032_at |
AT5G55860
|
unknown |
-0.339 |
-0.251 |
| 479 |
247046_at |
AT5G66540
|
unknown |
-0.339 |
-0.339 |
| 480 |
247859_at |
AT5G58410
|
[HEAT/U-box domain-containing protein] |
-0.338 |
0.027 |
| 481 |
244978_at |
ATCG00740
|
RPOA, RNA polymerase subunit alpha |
-0.337 |
-0.147 |
| 482 |
252546_at |
AT3G45830
|
unknown |
-0.336 |
-0.108 |
| 483 |
267132_at |
AT2G23420
|
NAPRT2, nicotinate phosphoribosyltransferase 2 |
-0.336 |
0.009 |
| 484 |
244979_at |
ATCG00750
|
RPS11, ribosomal protein S11 |
-0.335 |
-0.187 |
| 485 |
249521_at |
AT5G38690
|
[Zinc-finger domain of monoamine-oxidase A repressor R1 protein] |
-0.335 |
-0.219 |
| 486 |
246946_at |
AT5G25070
|
unknown |
-0.334 |
-0.209 |
| 487 |
266063_at |
AT2G18760
|
CHR8, chromatin remodeling 8 |
-0.334 |
-0.052 |
| 488 |
260598_at |
AT1G55930
|
[CBS domain-containing protein / transporter associated domain-containing protein] |
-0.334 |
-0.134 |
| 489 |
267570_at |
AT2G30800
|
ATVT-1, HVT1, helicase in vascular tissue and tapetum |
-0.334 |
-0.198 |
| 490 |
258151_at |
AT3G18080
|
BGLU44, B-S glucosidase 44 |
-0.334 |
0.046 |
| 491 |
257636_at |
AT3G26200
|
CYP71B22, cytochrome P450, family 71, subfamily B, polypeptide 22 |
-0.332 |
0.027 |
| 492 |
256623_at |
AT3G19960
|
ATM1, myosin 1 |
-0.332 |
-0.173 |
| 493 |
255836_at |
AT2G33440
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.332 |
-0.167 |
| 494 |
259638_at |
AT1G52360
|
[Coatomer, beta' subunit] |
-0.331 |
-0.089 |
| 495 |
250303_at |
AT5G12100
|
[pentatricopeptide (PPR) repeat-containing protein] |
-0.330 |
0.003 |
| 496 |
262137_at |
AT1G77920
|
TGA7, TGACG sequence-specific binding protein 7 |
-0.329 |
-0.198 |
| 497 |
257447_at |
AT2G04230
|
[FBD, F-box and Leucine Rich Repeat domains containing protein] |
-0.328 |
0.012 |
| 498 |
248099_at |
AT5G55300
|
MGO1, MGOUN 1, TOP1, TOPOISOMERASE 1, TOP1ALPHA, DNA topoisomerase I alpha |
-0.328 |
-0.304 |
| 499 |
260961_at |
AT1G05960
|
[ARM repeat superfamily protein] |
-0.327 |
-0.021 |
| 500 |
253468_at |
AT4G32160
|
[Phox (PX) domain-containing protein] |
-0.327 |
-0.176 |
| 501 |
253446_at |
AT4G32620
|
[Enhancer of polycomb-like transcription factor protein] |
-0.324 |
-0.187 |
| 502 |
258756_at |
AT3G11960
|
[Cleavage and polyadenylation specificity factor (CPSF) A subunit protein] |
-0.322 |
0.023 |
| 503 |
246555_at |
AT5G15470
|
GAUT14, galacturonosyltransferase 14 |
-0.322 |
-0.220 |
| 504 |
259285_at |
AT3G11460
|
MEF10, mitochondrial RNA editing factor 10 |
-0.321 |
-0.019 |
| 505 |
260592_at |
AT1G55850
|
ATCSLE1, CSLE1, cellulose synthase like E1 |
-0.321 |
0.025 |
| 506 |
262492_at |
AT1G21630
|
[Calcium-binding EF hand family protein] |
-0.321 |
0.041 |
| 507 |
256910_at |
AT3G24080
|
[KRR1 family protein] |
-0.320 |
-0.321 |
| 508 |
259698_at |
AT1G68930
|
[pentatricopeptide (PPR) repeat-containing protein] |
-0.320 |
-0.018 |
| 509 |
258573_at |
AT3G04260
|
PDE324, PIGMENT DEFECTIVE 324, PTAC3, plastid transcriptionally active 3 |
-0.319 |
-0.090 |
| 510 |
251629_at |
AT3G57410
|
ATVLN3, VLN3, villin 3 |
-0.318 |
0.038 |
| 511 |
262900_at |
AT1G59890
|
SNL5, SIN3-like 5 |
-0.318 |
-0.167 |
| 512 |
259117_at |
AT3G01320
|
SNL1, SIN3-like 1 |
-0.318 |
-0.272 |
| 513 |
267065_at |
AT2G41080
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.318 |
-0.087 |
| 514 |
255542_at |
AT4G01860
|
[Transducin family protein / WD-40 repeat family protein] |
-0.318 |
0.020 |
| 515 |
253092_at |
AT4G37380
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.318 |
-0.147 |
| 516 |
258043_at |
AT3G21290
|
[dentin sialophosphoprotein-related] |
-0.317 |
-0.206 |
| 517 |
255410_at |
AT4G03100
|
[Rho GTPase activating protein with PAK-box/P21-Rho-binding domain] |
-0.316 |
-0.229 |
| 518 |
261531_at |
AT1G63490
|
[transcription factor jumonji (jmjC) domain-containing protein] |
-0.316 |
-0.078 |
| 519 |
247443_at |
AT5G62720
|
[Integral membrane HPP family protein] |
-0.315 |
0.042 |
| 520 |
247874_at |
AT5G57710
|
SMAX1, SUPPRESSOR OF MAX2 1 |
-0.315 |
-0.060 |
| 521 |
265800_at |
AT2G35630
|
GEM1, MOR1, MICROTUBULE ORGANIZATION 1 |
-0.315 |
-0.029 |
| 522 |
258829_at |
AT3G07100
|
AtSEC24A, ERMO2, ENDOPLASMIC RETICULUM MORPHOLOGY 2, SEC24A |
-0.314 |
0.085 |
| 523 |
246568_at |
AT5G14960
|
DEL2, DP-E2F-like 2, E2FD, E2L1 |
-0.314 |
-0.039 |
| 524 |
250987_at |
AT5G02860
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.314 |
-0.035 |
| 525 |
260695_at |
AT1G32490
|
EMB2733, EMBRYO DEFECTIVE 2733, ESP3, ENHANCED SILENCING PHENOTYPE 3 |
-0.313 |
-0.171 |
| 526 |
258084_at |
AT3G26020
|
[Protein phosphatase 2A regulatory B subunit family protein] |
-0.313 |
-0.185 |
| 527 |
257971_at |
AT3G27530
|
GC6, golgin candidate 6, MAG4, MAIGO 4 |
-0.313 |
-0.158 |
| 528 |
247161_at |
AT5G65810
|
CGR3, cotton Golgi-related 3 |
-0.312 |
-0.018 |
| 529 |
256825_at |
AT3G22120
|
CWLP, cell wall-plasma membrane linker protein |
-0.312 |
-0.001 |
| 530 |
250267_at |
AT5G12930
|
unknown |
-0.312 |
0.044 |
| 531 |
264113_at |
AT2G31260
|
APG9, autophagy 9, ATAPG9 |
-0.312 |
0.038 |
| 532 |
255444_at |
AT4G02560
|
LD, luminidependens |
-0.312 |
-0.294 |
| 533 |
262996_at |
AT1G54440
|
[Polynucleotidyl transferase, ribonuclease H fold protein with HRDC domain] |
-0.312 |
-0.150 |
| 534 |
255762_at |
AT1G16710
|
HAC12, histone acetyltransferase of the CBP family 12 |
-0.311 |
-0.033 |
| 535 |
247049_at |
AT5G66440
|
unknown |
-0.311 |
0.047 |
| 536 |
263182_at |
AT1G05575
|
unknown |
-0.311 |
-0.288 |
| 537 |
248974_at |
AT5G45060
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.311 |
-0.100 |
| 538 |
246583_at |
AT5G14720
|
[Protein kinase superfamily protein] |
-0.310 |
-0.146 |
| 539 |
258529_at |
AT3G06740
|
GATA15, GATA transcription factor 15 |
-0.310 |
-0.069 |
| 540 |
256325_at |
AT3G02330
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.310 |
-0.027 |
| 541 |
252736_at |
AT3G43210
|
ATNACK2, ARABIDOPSIS NPK1-ACTIVATING KINESIN 2, NACK2, NPK1-ACTIVATING KINESIN 2, TES, TETRASPORE |
-0.309 |
-0.103 |
| 542 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
-0.309 |
-0.252 |
| 543 |
260823_at |
AT1G06770
|
DRIP1, DREB2A-interacting protein 1 |
-0.309 |
-0.104 |
| 544 |
251669_at |
AT3G57180
|
BPG2, BRASSINAZOLE(BRZ) INSENSITIVE PALE GREEN 2 |
-0.309 |
-0.089 |
| 545 |
251808_at |
AT3G55480
|
AP-3 beta, beta-subunit of adaptor protein complex 3, PAT2, protein affected trafficking 2, WAT1, WEAK ACID TOLERANT 1 |
-0.308 |
0.018 |
| 546 |
263112_at |
AT1G03080
|
NET1D, Networked 1D |
-0.308 |
-0.334 |
| 547 |
258601_at |
AT3G02760
|
[Class II aaRS and biotin synthetases superfamily protein] |
-0.307 |
-0.236 |
| 548 |
264384_at |
AT2G25170
|
CHD3, CHR6, CKH2, CYTOKININ-HYPERSENSITIVE 2, EPP1, ENHANCED PHOTOMORPHOGENIC 1, GYM, GYMNOS, PKL, PICKLE, SSL2, SUPPRESSOR OF SLR 2 |
-0.306 |
-0.132 |
| 549 |
267519_at |
AT2G30470
|
HSI2, high-level expression of sugar-inducible gene 2, VAL1, VP1/ABI3-LIKE 1 |
-0.306 |
-0.071 |
| 550 |
247004_at |
AT5G67570
|
DG1, DELAYED GREENING 1, EMB1408, embryo defective 1408, EMB246, EMBRYO DEFECTIVE 246 |
-0.306 |
-0.126 |
| 551 |
258818_at |
AT3G04580
|
EIN4, ETHYLENE INSENSITIVE 4 |
-0.306 |
-0.086 |
| 552 |
267009_at |
AT2G39260
|
[binding] |
-0.305 |
-0.186 |
| 553 |
250508_at |
AT5G09950
|
MEF7, mitochondrial editing factor 7 |
-0.305 |
0.001 |
| 554 |
245587_at |
AT4G15020
|
[hAT transposon superfamily] |
-0.305 |
-0.015 |
| 555 |
264472_at |
AT1G67140
|
SWEETIE, SWEETIE |
-0.304 |
-0.076 |
| 556 |
245182_at |
AT5G12430
|
DJC31, DNA J protein C31, TPR16, tetratricopeptide repeat 16 |
-0.303 |
-0.108 |
| 557 |
249266_at |
AT5G41670
|
[6-phosphogluconate dehydrogenase family protein] |
-0.302 |
-0.020 |
| 558 |
254140_at |
AT4G24610
|
unknown |
-0.302 |
-0.093 |
| 559 |
259672_at |
AT1G68990
|
MGP3, male gametophyte defective 3 |
-0.301 |
-0.096 |
| 560 |
248597_at |
AT5G49160
|
DDM2, DECREASED DNA METHYLATION 2, DMT01, DNA METHYLTRANSFERASE 01, DMT1, DNA METHYLTRANSFERASE 1, MET1, methyltransferase 1, MET2, METHYLTRANSFERASE 2, METI, METHYLTRANSFERASE I |
-0.301 |
0.017 |
| 561 |
263751_at |
AT2G21300
|
[ATP binding microtubule motor family protein] |
-0.300 |
-0.071 |
| 562 |
265536_at |
AT2G15880
|
[Leucine-rich repeat (LRR) family protein] |
-0.300 |
0.131 |
| 563 |
255409_at |
AT4G03090
|
AtNDX, NDX, Nodulin Homeobox |
-0.299 |
-0.157 |
| 564 |
252830_at |
AT4G39850
|
ABCD1, ATP-binding cassette D1, ACN2, acetate non-utilizing 2, AtABCD1, Arabidopsis thaliana ATP-binding cassette D1, CTS, COMATOSE, PED3, PEROXISOME DEFECTIVE 3, PXA1, peroxisomal ABC transporter 1 |
-0.299 |
0.009 |
| 565 |
261523_at |
AT1G71860
|
ATPTP1, PTP1, protein tyrosine phosphatase 1 |
-0.299 |
0.015 |
| 566 |
247841_at |
AT5G58040
|
ATFIP1[V], homolog of yeast FIP1 [V], ATFIPS5, FIP1[V], homolog of yeast FIP1 [V], FIPS5 |
-0.298 |
-0.234 |
| 567 |
256853_at |
AT3G18640
|
[Zinc finger C-x8-C-x5-C-x3-H type family protein] |
-0.298 |
-0.315 |
| 568 |
251675_at |
AT3G57300
|
ATINO80, INO80 ORTHOLOG, INO80, INO80 ortholog |
-0.298 |
-0.081 |
| 569 |
250787_at |
AT5G05560
|
APC1, anaphase promoting complex 1, EMB2771, EMBRYO DEFECTIVE 2771 |
-0.296 |
0.100 |
| 570 |
261551_at |
AT1G63440
|
HMA5, heavy metal atpase 5 |
-0.296 |
-0.018 |
| 571 |
244982_at |
ATCG00780
|
RPL14, ribosomal protein L14 |
-0.295 |
-0.406 |
| 572 |
258355_at |
AT3G14330
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.295 |
-0.116 |
| 573 |
260780_at |
AT1G14610
|
TWN2, TWIN 2, VALRS, VALYL TRNA SYNTHETASE |
-0.295 |
-0.216 |
| 574 |
254966_at |
AT4G11110
|
SPA2, SPA1-related 2 |
-0.294 |
-0.032 |
| 575 |
247228_at |
AT5G65140
|
TPPJ, trehalose-6-phosphate phosphatase J |
-0.294 |
0.015 |
| 576 |
266892_at |
AT2G26080
|
AtGLDP2, glycine decarboxylase P-protein 2, GLDP2, glycine decarboxylase P-protein 2 |
-0.293 |
-0.012 |
| 577 |
266727_at |
AT2G03150
|
emb1579, embryo defective 1579 |
-0.293 |
-0.309 |
| 578 |
248070_at |
AT5G55660
|
[DEK domain-containing chromatin associated protein] |
-0.293 |
-0.168 |
| 579 |
246232_at |
AT4G36630
|
EMB2754, EMBRYO DEFECTIVE 2754 |
-0.293 |
-0.036 |
| 580 |
247564_at |
AT5G61140
|
[U5 small nuclear ribonucleoprotein helicase] |
-0.291 |
-0.029 |
| 581 |
249326_at |
AT5G40840
|
AtRAD21.1, Sister chromatid cohesion 1 (SCC1) protein homolog 2, SYN2 |
-0.289 |
-0.094 |
| 582 |
253659_at |
AT4G30150
|
unknown |
-0.289 |
-0.052 |
| 583 |
244985_at |
ATCG00810
|
RPL22, ribosomal protein L22 |
-0.289 |
-0.386 |
| 584 |
250273_at |
AT5G13010
|
EMB3011, embryo defective 3011 |
-0.287 |
-0.161 |
| 585 |
264819_at |
AT1G03510
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.287 |
-0.088 |
| 586 |
259278_at |
AT3G01160
|
unknown |
-0.287 |
-0.330 |
| 587 |
258292_at |
AT3G23330
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.285 |
-0.121 |
| 588 |
245954_at |
AT5G28530
|
FRS10, FAR1-related sequence 10 |
-0.285 |
0.020 |
| 589 |
258361_at |
AT3G14270
|
FAB1B, FORMS APLOID AND BINUCLEATE CELLS 1B |
-0.285 |
-0.036 |
| 590 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
-0.284 |
0.079 |
| 591 |
252362_at |
AT3G48500
|
PDE312, PIGMENT DEFECTIVE 312, PTAC10, PLASTID TRANSCRIPTIONALLY ACTIVE 10, TAC10 |
-0.284 |
-0.285 |
| 592 |
248036_at |
AT5G55920
|
OLI2, OLIGOCELLULA 2 |
-0.283 |
-0.322 |
| 593 |
250569_at |
AT5G08130
|
BIM1 |
-0.283 |
-0.084 |
| 594 |
253605_at |
AT4G30990
|
[ARM repeat superfamily protein] |
-0.282 |
-0.266 |
| 595 |
250686_at |
AT5G06680
|
ATGCP3, ARABIDOPSIS THALIANA GAMMA TUBULIN COMPLEX PROTEIN 3, ATSPC98, SPINDLE POLE BODY COMPONENT 98, GCP3, GAMMA TUBULIN COMPLEX PROTEIN 3, SPC98, spindle pole body component 98 |
-0.282 |
0.003 |
| 596 |
264756_at |
AT1G61370
|
[S-locus lectin protein kinase family protein] |
-0.280 |
-0.100 |
| 597 |
248075_at |
AT5G55740
|
CRR21, chlororespiratory reduction 21 |
-0.279 |
-0.239 |
| 598 |
251128_at |
AT5G01110
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.279 |
-0.068 |
| 599 |
253649_at |
AT4G29790
|
unknown |
-0.279 |
-0.059 |
| 600 |
265225_at |
AT2G36720
|
[Acyl-CoA N-acyltransferase with RING/FYVE/PHD-type zinc finger domain] |
-0.278 |
0.036 |