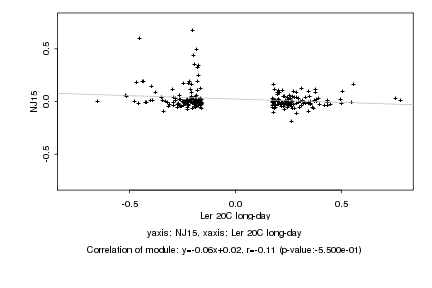
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Ler 20C long-day |
Log2 signal ratio
NJ15 |
| 1 |
247921_at |
AT5G57660
|
ATCOL5, BBX6, B-box domain protein 6, COL5, CONSTANS-like 5 |
0.778 |
0.012 |
| 2 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
0.752 |
0.037 |
| 3 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
0.554 |
0.166 |
| 4 |
247714_at |
AT5G59340
|
WOX2, WUSCHEL related homeobox 2 |
0.544 |
-0.009 |
| 5 |
247228_at |
AT5G65140
|
TPPJ, trehalose-6-phosphate phosphatase J |
0.505 |
0.097 |
| 6 |
246868_at |
AT5G26010
|
[Protein phosphatase 2C family protein] |
0.501 |
-0.018 |
| 7 |
253009_at |
AT4G37930
|
SHM1, serine transhydroxymethyltransferase 1, SHMT1, SERINE HYDROXYMETHYLTRANSFERASE 1, STM, SERINE TRANSHYDROXYMETHYLTRANSFERASE |
0.496 |
0.023 |
| 8 |
248139_at |
AT5G54970
|
unknown |
0.449 |
-0.027 |
| 9 |
247151_at |
AT5G65640
|
bHLH093, beta HLH protein 93 |
0.434 |
0.000 |
| 10 |
258471_at |
AT3G06030
|
ANP3, NPK1-related protein kinase 3, AtANP3, MAPKKK12, NP3, NPK1-related protein kinase 3 |
0.434 |
-0.035 |
| 11 |
245803_at |
AT1G47128
|
RD21, responsive to dehydration 21, RD21A, responsive to dehydration 21A |
0.431 |
0.019 |
| 12 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.420 |
-0.032 |
| 13 |
255410_at |
AT4G03100
|
[Rho GTPase activating protein with PAK-box/P21-Rho-binding domain] |
0.397 |
-0.023 |
| 14 |
245585_at |
AT4G14970
|
unknown |
0.388 |
0.035 |
| 15 |
261749_at |
AT1G76180
|
ERD14, EARLY RESPONSE TO DEHYDRATION 14 |
0.380 |
0.002 |
| 16 |
252234_at |
AT3G49780
|
ATPSK3 (FORMER SYMBOL), ATPSK4, phytosulfokine 4 precursor, PSK4, phytosulfokine 4 precursor |
0.379 |
0.025 |
| 17 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.376 |
0.116 |
| 18 |
250292_at |
AT5G13220
|
JAS1, JASMONATE-ASSOCIATED 1, JAZ10, jasmonate-zim-domain protein 10, TIFY9, TIFY DOMAIN PROTEIN 9 |
0.375 |
0.088 |
| 19 |
245138_at |
AT2G45190
|
AFO, ABNORMAL FLORAL ORGANS, FIL, FILAMENTOUS FLOWER, YAB1, YABBY1 |
0.372 |
0.010 |
| 20 |
248551_at |
AT5G50200
|
ATNRT3.1, NRT3.1, NITRATE TRANSPORTER 3.1, WR3, WOUND-RESPONSIVE 3 |
0.370 |
0.001 |
| 21 |
256332_at |
AT1G76890
|
AT-GT2, GT2 |
0.368 |
-0.059 |
| 22 |
251144_at |
AT5G01210
|
[HXXXD-type acyl-transferase family protein] |
0.364 |
-0.052 |
| 23 |
254166_at |
AT4G24190
|
AtHsp90-7, HEAT SHOCK PROTEIN 90-7, AtHsp90.7, HEAT SHOCK PROTEIN 90.7, HSP90.7, HEAT SHOCK PROTEIN 90.7, SHD, SHEPHERD |
0.353 |
0.009 |
| 24 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.352 |
-0.023 |
| 25 |
265475_at |
AT2G15620
|
ATHNIR, ARABIDOPSIS THALIANA NITRITE REDUCTASE, NIR, NITRITE REDUCTASE, NIR1, nitrite reductase 1 |
0.351 |
-0.016 |
| 26 |
260774_at |
AT1G78290
|
SNRK2-8, SNF1-RELATED PROTEIN KINASE 2-8, SNRK2.8, SNF1-RELATED PROTEIN KINASE 2.8, SRK2C, SNF1-RELATED PROTEIN KINASE 2C |
0.346 |
0.048 |
| 27 |
255590_at |
AT4G01610
|
[Cysteine proteinases superfamily protein] |
0.345 |
-0.004 |
| 28 |
257636_at |
AT3G26200
|
CYP71B22, cytochrome P450, family 71, subfamily B, polypeptide 22 |
0.342 |
-0.088 |
| 29 |
267028_at |
AT2G38470
|
ATWRKY33, WRKY DNA-BINDING PROTEIN 33, WRKY33, WRKY DNA-binding protein 33 |
0.341 |
0.101 |
| 30 |
262284_at |
AT1G68670
|
[myb-like transcription factor family protein] |
0.341 |
-0.014 |
| 31 |
245987_at |
AT5G13180
|
ANAC083, NAC domain containing protein 83, NAC083, NAC domain containing protein 83, VNI2, VND-interacting 2 |
0.331 |
-0.018 |
| 32 |
251787_at |
AT3G55410
|
[2-oxoglutarate dehydrogenase, E1 component] |
0.330 |
0.041 |
| 33 |
247867_at |
AT5G57630
|
CIPK21, CBL-interacting protein kinase 21, SnRK3.4, SNF1-RELATED PROTEIN KINASE 3.4 |
0.325 |
-0.008 |
| 34 |
256825_at |
AT3G22120
|
CWLP, cell wall-plasma membrane linker protein |
0.321 |
-0.015 |
| 35 |
258833_at |
AT3G07274
|
|
0.313 |
0.011 |
| 36 |
245041_at |
AT2G26530
|
AR781 |
0.312 |
0.125 |
| 37 |
249020_at |
AT5G44800
|
CHR4, chromatin remodeling 4, PKR1, PICKLE RELATED 1 |
0.312 |
-0.036 |
| 38 |
257867_at |
AT3G17780
|
unknown |
0.307 |
0.009 |
| 39 |
266009_at |
AT2G37420
|
[ATP binding microtubule motor family protein] |
0.302 |
-0.054 |
| 40 |
248756_at |
AT5G47560
|
ATSDAT, ATTDT, TONOPLAST DICARBOXYLATE TRANSPORTER, TDT, tonoplast dicarboxylate transporter |
0.296 |
-0.009 |
| 41 |
266447_at |
AT2G43290
|
MSS3, multicopy suppressors of snf4 deficiency in yeast 3 |
0.295 |
0.031 |
| 42 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.287 |
0.045 |
| 43 |
254652_at |
AT4G18170
|
ATWRKY28, WRKY28, WRKY DNA-binding protein 28 |
0.287 |
-0.017 |
| 44 |
265539_at |
AT2G15830
|
unknown |
0.286 |
-0.111 |
| 45 |
263182_at |
AT1G05575
|
unknown |
0.284 |
0.090 |
| 46 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
0.278 |
-0.057 |
| 47 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.275 |
0.044 |
| 48 |
266461_at |
AT2G47730
|
ATGSTF5, GLUTATHIONE S-TRANSFERASE (CLASS PHI) 5, ATGSTF8, Arabidopsis thaliana glutathione S-transferase phi 8, GST6, GSTF8, glutathione S-transferase phi 8 |
0.272 |
0.100 |
| 49 |
245302_at |
AT4G17695
|
KAN3, KANADI 3 |
0.271 |
-0.026 |
| 50 |
247085_at |
AT5G66310
|
[ATP binding microtubule motor family protein] |
0.269 |
-0.064 |
| 51 |
262180_at |
AT1G78050
|
PGM, phosphoglycerate/bisphosphoglycerate mutase |
0.269 |
0.004 |
| 52 |
249426_at |
AT5G39840
|
[ATP-dependent RNA helicase, mitochondrial, putative] |
0.265 |
0.051 |
| 53 |
247162_at |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
0.264 |
-0.187 |
| 54 |
257540_at |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
0.263 |
0.001 |
| 55 |
261410_at |
AT1G07610
|
MT1C, metallothionein 1C |
0.261 |
-0.038 |
| 56 |
260267_at |
AT1G68530
|
CER6, ECERIFERUM 6, CUT1, CUTICULAR 1, G2, KCS6, 3-ketoacyl-CoA synthase 6, POP1, POLLEN-PISTIL INCOMPATIBILITY 1 |
0.259 |
0.036 |
| 57 |
251946_at |
AT3G53540
|
TRM19, TON1 Recruiting Motif 19 |
0.256 |
-0.021 |
| 58 |
259679_at |
AT1G77720
|
PPK1, putative protein kinase 1 |
0.256 |
-0.014 |
| 59 |
249266_at |
AT5G41670
|
[6-phosphogluconate dehydrogenase family protein] |
0.256 |
-0.025 |
| 60 |
259516_at |
AT1G20450
|
ERD10, EARLY RESPONSIVE TO DEHYDRATION 10, LTI29, LOW TEMPERATURE INDUCED 29, LTI45, LOW TEMPERATURE INDUCED 45 |
0.252 |
0.064 |
| 61 |
266063_at |
AT2G18760
|
CHR8, chromatin remodeling 8 |
0.249 |
0.028 |
| 62 |
259681_at |
AT1G77760
|
GNR1, NIA1, nitrate reductase 1, NR1, NITRATE REDUCTASE 1 |
0.249 |
-0.007 |
| 63 |
247049_at |
AT5G66440
|
unknown |
0.249 |
-0.036 |
| 64 |
254083_at |
AT4G24920
|
[secE/sec61-gamma protein transport protein] |
0.247 |
-0.000 |
| 65 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
0.245 |
-0.001 |
| 66 |
248200_at |
AT5G54160
|
ATOMT1, O-methyltransferase 1, COMT1, caffeate O-methyltransferase 1, OMT1, O-methyltransferase 1 |
0.244 |
-0.002 |
| 67 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
0.244 |
0.046 |
| 68 |
266778_at |
AT2G29090
|
CYP707A2, cytochrome P450, family 707, subfamily A, polypeptide 2 |
0.244 |
-0.035 |
| 69 |
264690_at |
AT1G70060
|
SNL4, SIN3-like 4 |
0.243 |
-0.053 |
| 70 |
250916_at |
AT5G03630
|
ATMDAR2 |
0.242 |
0.003 |
| 71 |
256351_at |
AT1G54960
|
ANP2, NPK1-related protein kinase 2, MAPKKK2, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASES 2, NP2, NPK1-related protein kinase 2 |
0.240 |
0.003 |
| 72 |
246261_at |
AT1G31810
|
AFH14, Formin Homology 14 |
0.240 |
-0.015 |
| 73 |
263799_at |
AT2G24550
|
unknown |
0.236 |
0.003 |
| 74 |
253203_at |
AT4G34710
|
ADC2, arginine decarboxylase 2, ATADC2, SPE2 |
0.232 |
0.016 |
| 75 |
247980_at |
AT5G56860
|
GATA21, GATA TRANSCRIPTION FACTOR 21, GNC, GATA, nitrate-inducible, carbon metabolism-involved |
0.231 |
-0.024 |
| 76 |
255542_at |
AT4G01860
|
[Transducin family protein / WD-40 repeat family protein] |
0.230 |
-0.067 |
| 77 |
264948_at |
AT1G77030
|
[hydrolases, acting on acid anhydrides, in phosphorus-containing anhydrides] |
0.229 |
0.053 |
| 78 |
245977_at |
AT5G13110
|
G6PD2, glucose-6-phosphate dehydrogenase 2 |
0.224 |
-0.026 |
| 79 |
248070_at |
AT5G55660
|
[DEK domain-containing chromatin associated protein] |
0.224 |
-0.038 |
| 80 |
263963_at |
AT2G36080
|
ABS2, ABNORMAL SHOOT 2, NGAL1, NGATHA-Like 1 |
0.224 |
0.051 |
| 81 |
256623_at |
AT3G19960
|
ATM1, myosin 1 |
0.220 |
-0.005 |
| 82 |
248799_at |
AT5G47230
|
ATERF-5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR- 5, ATERF5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 5, AtMACD1, ERF102, ERF5, ethylene responsive element binding factor 5 |
0.220 |
0.112 |
| 83 |
250374_at |
AT5G11530
|
EMF1, embryonic flower 1 |
0.216 |
-0.005 |
| 84 |
262899_at |
AT1G59870
|
ABCG36, ATP-binding cassette G36, ATABCG36, Arabidopsis thaliana ATP-binding cassette G36, ATPDR8, ARABIDOPSIS PLEIOTROPIC DRUG RESISTANCE 8, PDR8, PLEIOTROPIC DRUG RESISTANCE 8, PEN3, PENETRATION 3 |
0.214 |
-0.026 |
| 85 |
259303_at |
AT3G05130
|
unknown |
0.214 |
-0.024 |
| 86 |
263937_at |
AT2G35910
|
[RING/U-box superfamily protein] |
0.213 |
-0.045 |
| 87 |
260031_at |
AT1G68790
|
CRWN3, CROWDED NUCLEI 3, LINC3, LITTLE NUCLEI3 |
0.211 |
-0.028 |
| 88 |
248841_at |
AT5G46740
|
UBP21, ubiquitin-specific protease 21 |
0.208 |
-0.007 |
| 89 |
245280_at |
AT4G16845
|
VRN2, REDUCED VERNALIZATION RESPONSE 2 |
0.206 |
0.014 |
| 90 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
0.205 |
0.092 |
| 91 |
250788_at |
AT5G05570
|
[transducin family protein / WD-40 repeat family protein] |
0.203 |
0.011 |
| 92 |
247443_at |
AT5G62720
|
[Integral membrane HPP family protein] |
0.203 |
0.021 |
| 93 |
253999_at |
AT4G26200
|
ACCS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ACS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ATACS7 |
0.203 |
0.109 |
| 94 |
261193_at |
AT1G32920
|
unknown |
0.202 |
0.087 |
| 95 |
246445_at |
AT5G17630
|
[Nucleotide/sugar transporter family protein] |
0.202 |
-0.024 |
| 96 |
264204_at |
AT1G22710
|
ATSUC2, ARABIDOPSIS THALIANA SUCROSE-PROTON SYMPORTER 2, SUC2, sucrose-proton symporter 2, SUT1, SUCROSE TRANSPORTER 1 |
0.202 |
0.026 |
| 97 |
254260_at |
AT4G23440
|
[Disease resistance protein (TIR-NBS class)] |
0.198 |
0.019 |
| 98 |
264246_at |
AT1G60140
|
ATTPS10, trehalose phosphate synthase, TPS10, trehalose phosphate synthase, TPS10, TREHALOSE PHOSPHATE SYNTHASE 10 |
0.197 |
-0.022 |
| 99 |
245259_at |
AT4G14150
|
KINESIN-12A, PAKRP1, phragmoplast-associated kinesin-related protein 1 |
0.197 |
-0.019 |
| 100 |
265672_at |
AT2G31980
|
AtCYS2, PHYTOCYSTATIN 2, CYS2, PHYTOCYSTATIN 2 |
0.196 |
0.068 |
| 101 |
247026_at |
AT5G67080
|
MAPKKK19, mitogen-activated protein kinase kinase kinase 19 |
0.196 |
0.013 |
| 102 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
0.196 |
0.088 |
| 103 |
256277_at |
AT3G12120
|
AtFAD2, FAD2, fatty acid desaturase 2 |
0.196 |
0.005 |
| 104 |
253133_at |
AT4G35800
|
NRPB1, RNA polymerase II large subunit, RNA_POL_II_LS, RNA POLYMERASE II LARGE SUBUNIT, RNA_POL_II_LSRNA_POL_II_LS, RPB1, RNA POLYMERASE II LARGE SUBUNIT |
0.189 |
-0.049 |
| 105 |
245763_at |
AT1G27850
|
unknown |
0.189 |
0.007 |
| 106 |
261378_at |
AT1G18890
|
ATCDPK1, calcium-dependent protein kinase 1, AtCPK10, CDPK1, calcium-dependent protein kinase 1, CPK10 |
0.188 |
0.007 |
| 107 |
266518_at |
AT2G35170
|
[Histone H3 K4-specific methyltransferase SET7/9 family protein] |
0.188 |
-0.031 |
| 108 |
259376_at |
AT3G16320
|
Tetratricopeptide repeat (TPR)-like superfamily protein |
0.185 |
0.003 |
| 109 |
250460_at |
AT5G09850
|
[Transcription elongation factor (TFIIS) family protein] |
0.185 |
0.012 |
| 110 |
253373_at |
AT4G33150
|
LKR, LKR/SDH, LYSINE-KETOGLUTARATE REDUCTASE/SACCHAROPINE DEHYDROGENASE, SDH, SACCHAROPINE DEHYDROGENASE |
0.185 |
-0.027 |
| 111 |
245317_at |
AT4G15610
|
[Uncharacterised protein family (UPF0497)] |
0.184 |
0.019 |
| 112 |
264424_at |
AT1G61740
|
[Sulfite exporter TauE/SafE family protein] |
0.183 |
0.009 |
| 113 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.183 |
0.116 |
| 114 |
249871_at |
AT5G23110
|
[Zinc finger, C3HC4 type (RING finger) family protein] |
0.183 |
-0.048 |
| 115 |
247149_at |
AT5G65660
|
[hydroxyproline-rich glycoprotein family protein] |
0.182 |
0.004 |
| 116 |
246583_at |
AT5G14720
|
[Protein kinase superfamily protein] |
0.182 |
-0.058 |
| 117 |
255605_at |
AT4G01090
|
unknown |
0.182 |
-0.037 |
| 118 |
261117_at |
AT1G75310
|
AUL1, auxilin-like 1 |
0.181 |
-0.033 |
| 119 |
245158_at |
AT2G33130
|
RALF18, RALFL18, ralf-like 18 |
0.181 |
0.015 |
| 120 |
246718_at |
AT5G28930
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT2G07240.1)] |
0.181 |
-0.023 |
| 121 |
266964_at |
AT2G39480
|
ABCB6, ATP-binding cassette B6, PGP6, P-glycoprotein 6 |
0.180 |
-0.034 |
| 122 |
267326_at |
AT2G19380
|
[RNA recognition motif (RRM)-containing protein] |
0.180 |
-0.029 |
| 123 |
266313_at |
AT2G26980
|
CIPK3, CBL-interacting protein kinase 3, SnRK3.17, SNF1-RELATED PROTEIN KINASE 3.17 |
0.177 |
0.021 |
| 124 |
254238_at |
AT4G23540
|
[ARM repeat superfamily protein] |
0.177 |
0.010 |
| 125 |
262001_at |
AT1G33790
|
[jacalin lectin family protein] |
0.176 |
-0.098 |
| 126 |
262362_at |
AT1G72840
|
[Disease resistance protein (TIR-NBS-LRR class)] |
0.176 |
-0.022 |
| 127 |
255900_at |
AT1G17830
|
unknown |
0.176 |
0.163 |
| 128 |
250937_at |
AT5G03230
|
unknown |
0.175 |
-0.001 |
| 129 |
264535_at |
AT1G55690
|
[Sec14p-like phosphatidylinositol transfer family protein] |
0.174 |
0.029 |
| 130 |
258458_at |
AT3G22430
|
unknown |
0.174 |
0.001 |
| 131 |
264473_at |
AT1G67180
|
[zinc finger (C3HC4-type RING finger) family protein / BRCT domain-containing protein] |
0.174 |
-0.034 |
| 132 |
250809_at |
AT5G05140
|
[Transcription elongation factor (TFIIS) family protein] |
0.171 |
-0.048 |
| 133 |
248194_at |
AT5G54095
|
unknown |
-0.655 |
0.001 |
| 134 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
-0.523 |
0.066 |
| 135 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
-0.519 |
0.050 |
| 136 |
257821_at |
AT3G25170
|
RALFL26, ralf-like 26 |
-0.481 |
0.005 |
| 137 |
255811_at |
AT4G10250
|
ATHSP22.0 |
-0.469 |
0.186 |
| 138 |
263144_at |
AT1G54070
|
[Dormancy/auxin associated family protein] |
-0.462 |
-0.018 |
| 139 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
-0.456 |
0.605 |
| 140 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
-0.441 |
0.195 |
| 141 |
254059_at |
AT4G25200
|
ATHSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6, HSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6 |
-0.435 |
0.195 |
| 142 |
254430_at |
AT4G20820
|
[FAD-binding Berberine family protein] |
-0.428 |
-0.004 |
| 143 |
251228_at |
AT3G62710
|
[Glycosyl hydrolase family protein] |
-0.421 |
-0.004 |
| 144 |
251590_at |
AT3G57690
|
AGP23, arabinogalactan protein 23, ATAGP23, ARABINOGALACTAN-PROTEIN 23 |
-0.402 |
0.015 |
| 145 |
254263_at |
AT4G23493
|
unknown |
-0.399 |
0.144 |
| 146 |
266764_at |
AT2G47050
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.394 |
0.011 |
| 147 |
264912_at |
AT1G60750
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.382 |
0.086 |
| 148 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
-0.351 |
0.044 |
| 149 |
250174_at |
AT5G14380
|
AGP6, arabinogalactan protein 6 |
-0.345 |
0.014 |
| 150 |
258971_at |
AT3G01990
|
ACR6, ACT domain repeat 6 |
-0.340 |
-0.091 |
| 151 |
258889_at |
AT3G05610
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.333 |
0.002 |
| 152 |
262760_at |
AT1G10770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.325 |
-0.006 |
| 153 |
256062_at |
AT1G07090
|
LSH6, LIGHT SENSITIVE HYPOCOTYLS 6 |
-0.318 |
-0.038 |
| 154 |
251180_at |
AT3G62640
|
unknown |
-0.315 |
-0.024 |
| 155 |
267389_at |
AT2G44460
|
BGLU28, beta glucosidase 28 |
-0.300 |
0.122 |
| 156 |
254583_at |
AT4G19480
|
unknown |
-0.296 |
-0.008 |
| 157 |
250022_at |
AT5G18210
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.296 |
-0.037 |
| 158 |
265022_at |
AT1G24520
|
BCP1, homolog of Brassica campestris pollen protein 1 |
-0.294 |
0.041 |
| 159 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-0.286 |
0.034 |
| 160 |
260907_at |
AT1G02575
|
unknown |
-0.284 |
0.015 |
| 161 |
257635_at |
AT3G26280
|
CYP71B4, cytochrome P450, family 71, subfamily B, polypeptide 4 |
-0.278 |
-0.020 |
| 162 |
260011_at |
AT1G68110
|
[ENTH/ANTH/VHS superfamily protein] |
-0.278 |
-0.052 |
| 163 |
251323_at |
AT3G61580
|
AtSLD1, SLD1, sphingoid LCB desaturase 1 |
-0.273 |
-0.019 |
| 164 |
266921_at |
AT2G45970
|
CYP86A8, cytochrome P450, family 86, subfamily A, polypeptide 8, LCR, LACERATA |
-0.270 |
-0.013 |
| 165 |
248534_at |
AT5G50030
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.268 |
0.021 |
| 166 |
260462_at |
AT1G10970
|
ATZIP4, ZIP4, zinc transporter 4 precursor |
-0.266 |
-0.033 |
| 167 |
262148_at |
AT1G52560
|
[HSP20-like chaperones superfamily protein] |
-0.265 |
0.060 |
| 168 |
247236_at |
AT5G64590
|
unknown |
-0.263 |
0.021 |
| 169 |
250631_at |
AT5G07430
|
[Pectin lyase-like superfamily protein] |
-0.263 |
-0.040 |
| 170 |
258077_at |
AT3G26110
|
[Anther-specific protein agp1-like] |
-0.258 |
-0.041 |
| 171 |
248822_at |
AT5G47000
|
[Peroxidase superfamily protein] |
-0.252 |
0.009 |
| 172 |
245243_at |
AT1G44414
|
unknown |
-0.250 |
-0.036 |
| 173 |
248389_at |
AT5G51990
|
CBF4, C-repeat-binding factor 4, DREB1D, DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN 1D |
-0.249 |
0.171 |
| 174 |
261198_at |
AT1G12940
|
ATNRT2.5, nitrate transporter2.5, NRT2.5, nitrate transporter2.5 |
-0.247 |
-0.002 |
| 175 |
265080_at |
AT1G55570
|
sks12, SKU5 similar 12 |
-0.246 |
0.002 |
| 176 |
259216_at |
AT3G09000
|
[proline-rich family protein] |
-0.245 |
-0.017 |
| 177 |
254024_at |
AT4G25780
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.244 |
0.009 |
| 178 |
263950_at |
AT2G36020
|
HVA22J, HVA22-like protein J |
-0.240 |
-0.017 |
| 179 |
267398_at |
AT2G44560
|
AtGH9B11, glycosyl hydrolase 9B11, GH9B11, glycosyl hydrolase 9B11 |
-0.237 |
-0.026 |
| 180 |
248753_at |
AT5G47630
|
mtACP3, mitochondrial acyl carrier protein 3 |
-0.235 |
0.015 |
| 181 |
255690_at |
AT4G00360
|
ATT1, ABERRANT INDUCTION OF TYPE THREE 1, CYP86A2, cytochrome P450, family 86, subfamily A, polypeptide 2 |
-0.233 |
-0.017 |
| 182 |
246540_at |
AT5G15600
|
SP1L4, SPIRAL1-like4 |
-0.230 |
-0.067 |
| 183 |
263124_at |
AT1G78480
|
[Prenyltransferase family protein] |
-0.229 |
-0.056 |
| 184 |
249548_at |
AT5G38170
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.229 |
0.027 |
| 185 |
258434_at |
AT3G16770
|
ATEBP, ethylene-responsive element binding protein, EBP, ethylene-responsive element binding protein, ERF72, ETHYLENE RESPONSE FACTOR 72, RAP2.3, RELATED TO AP2 3 |
-0.227 |
-0.007 |
| 186 |
246381_at |
AT1G39430
|
unknown |
-0.226 |
0.005 |
| 187 |
251166_at |
AT3G63350
|
AT-HSFA7B, HSFA7B, HEAT SHOCK TRANSCRIPTION FACTOR A7B |
-0.225 |
0.173 |
| 188 |
254457_at |
AT4G21170
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.223 |
-0.027 |
| 189 |
260170_at |
AT1G71890
|
ATSUC5, SUCROSE-PROTON SYMPORTER 5, SUC5 |
-0.223 |
0.002 |
| 190 |
263726_at |
AT2G13610
|
ABCG5, ATP-binding cassette G5 |
-0.222 |
-0.036 |
| 191 |
252036_at |
AT3G52070
|
unknown |
-0.222 |
-0.001 |
| 192 |
263875_at |
AT2G21970
|
SEP2, stress enhanced protein 2 |
-0.222 |
-0.005 |
| 193 |
258336_at |
AT3G16050
|
A37, ATPDX1.2, ARABIDOPSIS THALIANA PYRIDOXINE BIOSYNTHESIS 1.2, PDX1.2, pyridoxine biosynthesis 1.2 |
-0.221 |
0.194 |
| 194 |
247774_at |
AT5G58660
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.219 |
0.003 |
| 195 |
260978_at |
AT1G53540
|
[HSP20-like chaperones superfamily protein] |
-0.215 |
0.120 |
| 196 |
249581_at |
AT5G37600
|
ATGLN1;1, ARABIDOPSIS GLUTAMINE SYNTHASE 1;1, ATGSR1, ARABIDOPSIS THALIANA GLUTAMINE SYNTHASE CLONE R1, GLN1;1, GLUTAMINE SYNTHASE 1;1, GSR 1, glutamine synthase clone R1 |
-0.215 |
0.011 |
| 197 |
255760_at |
AT1G16780
|
AtVHP2;2, VHP2;2 |
-0.212 |
-0.002 |
| 198 |
262098_at |
AT1G56170
|
ATHAP5B, HAP5B, NF-YC2, nuclear factor Y, subunit C2 |
-0.211 |
0.166 |
| 199 |
258827_at |
AT3G07150
|
unknown |
-0.211 |
0.092 |
| 200 |
251060_at |
AT5G01820
|
ATCIPK14, ATSR1, serine/threonine protein kinase 1, CIPK14, CBL-INTERACTING PROTEIN KINASE 14, PKS24, SOS2-like protein kinase 24, SnRK3.15, SNF1-RELATED PROTEIN KINASE 3.15, SR1, serine/threonine protein kinase 1 |
-0.210 |
0.057 |
| 201 |
247126_at |
AT5G66080
|
APD9, Arabidopsis Pp2c clade D 9 |
-0.210 |
0.024 |
| 202 |
248367_at |
AT5G52360
|
ADF10, actin depolymerizing factor 10 |
-0.209 |
0.044 |
| 203 |
252052_at |
AT3G52600
|
AtCWIN2, AtcwINV2, cell wall invertase 2, CWINV2, cell wall invertase 2 |
-0.208 |
0.035 |
| 204 |
259160_at |
AT3G05410
|
[Photosystem II reaction center PsbP family protein] |
-0.208 |
0.039 |
| 205 |
248137_at |
AT5G54950
|
[Aconitase family protein] |
-0.208 |
0.027 |
| 206 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
-0.206 |
0.683 |
| 207 |
265847_at |
AT2G35750
|
unknown |
-0.204 |
-0.058 |
| 208 |
262870_at |
AT1G64710
|
[GroES-like zinc-binding dehydrogenase family protein] |
-0.203 |
-0.011 |
| 209 |
245367_at |
AT4G16265
|
NRPB9B, NRPD9B, NRPE9B |
-0.203 |
-0.012 |
| 210 |
245548_at |
AT4G15310
|
CYP702A3, cytochrome P450, family 702, subfamily A, polypeptide 3 |
-0.202 |
0.027 |
| 211 |
258111_at |
AT3G14630
|
CYP72A9, cytochrome P450, family 72, subfamily A, polypeptide 9 |
-0.202 |
-0.006 |
| 212 |
261294_at |
AT1G48430
|
[Dihydroxyacetone kinase] |
-0.201 |
-0.015 |
| 213 |
249575_at |
AT5G37670
|
[HSP20-like chaperones superfamily protein] |
-0.199 |
0.440 |
| 214 |
261804_at |
AT1G30530
|
UGT78D1, UDP-glucosyl transferase 78D1 |
-0.198 |
-0.045 |
| 215 |
258339_at |
AT3G16120
|
[Dynein light chain type 1 family protein] |
-0.198 |
-0.007 |
| 216 |
261032_at |
AT1G17430
|
[alpha/beta-Hydrolases superfamily protein] |
-0.195 |
0.010 |
| 217 |
256518_at |
AT1G66080
|
unknown |
-0.194 |
0.358 |
| 218 |
245201_at |
AT1G67840
|
CSK, chloroplast sensor kinase |
-0.193 |
0.008 |
| 219 |
254671_at |
AT4G18410
|
[Mutator-like transposase family, has a 8.7e-30 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
-0.191 |
-0.006 |
| 220 |
247897_at |
AT5G57810
|
TET15, tetraspanin15 |
-0.191 |
-0.019 |
| 221 |
250281_at |
AT5G13240
|
[transcription regulators] |
-0.191 |
-0.028 |
| 222 |
263668_at |
AT1G04350
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.190 |
-0.049 |
| 223 |
247214_at |
AT5G64850
|
unknown |
-0.189 |
0.027 |
| 224 |
251896_at |
AT3G54390
|
[sequence-specific DNA binding transcription factors] |
-0.189 |
0.049 |
| 225 |
249203_at |
AT5G42590
|
CYP71A16, cytochrome P450, family 71, subfamily A, polypeptide 16, MRO, marneral oxidase |
-0.188 |
0.060 |
| 226 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
-0.187 |
0.496 |
| 227 |
262546_at |
AT1G31260
|
ZIP10, zinc transporter 10 precursor |
-0.186 |
0.019 |
| 228 |
251358_at |
AT3G61160
|
[Protein kinase superfamily protein] |
-0.186 |
-0.018 |
| 229 |
255357_at |
AT4G03930
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.185 |
0.021 |
| 230 |
249479_at |
AT5G38960
|
[RmlC-like cupins superfamily protein] |
-0.185 |
-0.043 |
| 231 |
267001_at |
AT2G34470
|
PSKF109, UREG, urease accessory protein G |
-0.183 |
-0.009 |
| 232 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
-0.182 |
0.199 |
| 233 |
261994_at |
AT1G33640
|
unknown |
-0.181 |
-0.001 |
| 234 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
-0.181 |
0.326 |
| 235 |
247964_at |
AT5G56600
|
PFN3, PROFILIN 3, PRF3, profilin 3 |
-0.181 |
0.107 |
| 236 |
259429_at |
AT1G01600
|
CYP86A4, cytochrome P450, family 86, subfamily A, polypeptide 4 |
-0.180 |
-0.005 |
| 237 |
254424_at |
AT4G21510
|
AtFBS2, FBS2, F-box stress induced 2 |
-0.179 |
-0.006 |
| 238 |
249375_at |
AT5G40730
|
AGP24, arabinogalactan protein 24, ATAGP24, ARABIDOPSIS THALIANA ARABINOGALACTAN PROTEIN 24 |
-0.178 |
0.020 |
| 239 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
-0.177 |
0.254 |
| 240 |
246464_at |
AT5G16980
|
[Zinc-binding dehydrogenase family protein] |
-0.176 |
0.347 |
| 241 |
266579_at |
AT2G23930
|
SNRNP-G, probable small nuclear ribonucleoprotein G |
-0.175 |
-0.030 |
| 242 |
253181_at |
AT4G35180
|
LHT7, LYS/HIS transporter 7 |
-0.174 |
-0.018 |
| 243 |
250945_at |
AT5G03400
|
unknown |
-0.173 |
0.013 |
| 244 |
254280_at |
AT4G22756
|
ATSMO1-2, SMO1-2, sterol C4-methyl oxidase 1-2 |
-0.173 |
-0.005 |
| 245 |
267535_at |
AT2G41940
|
ZFP8, zinc finger protein 8 |
-0.170 |
-0.018 |
| 246 |
254630_at |
AT4G18360
|
GOX3, glycolate oxidase 3 |
-0.170 |
0.025 |
| 247 |
245057_at |
AT2G26490
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.170 |
-0.015 |
| 248 |
251391_at |
AT3G60910
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.170 |
-0.009 |
| 249 |
251644_at |
AT3G57540
|
[Remorin family protein] |
-0.169 |
0.132 |
| 250 |
256943_at |
AT3G18910
|
ETP2, EIN2 targeting protein2 |
-0.168 |
-0.024 |
| 251 |
254360_at |
AT4G22340
|
CDS2, cytidinediphosphate diacylglycerol synthase 2 |
-0.168 |
0.029 |
| 252 |
254239_at |
AT4G23400
|
PIP1;5, plasma membrane intrinsic protein 1;5, PIP1D |
-0.167 |
-0.030 |
| 253 |
248946_at |
AT5G45530
|
unknown |
-0.167 |
-0.014 |
| 254 |
258962_at |
AT3G10570
|
CYP77A6, cytochrome P450, family 77, subfamily A, polypeptide 6 |
-0.166 |
-0.055 |
| 255 |
247295_at |
AT5G64180
|
unknown |
-0.166 |
0.013 |
| 256 |
250477_at |
AT5G10190
|
[Major facilitator superfamily protein] |
-0.166 |
0.006 |
| 257 |
265833_at |
AT2G14420
|
[Mutator-like transposase family, has a 8.4e-10 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
-0.165 |
-0.017 |
| 258 |
245386_at |
AT4G14010
|
RALFL32, ralf-like 32 |
-0.165 |
-0.007 |
| 259 |
262198_at |
AT1G53830
|
ATPME2, pectin methylesterase 2, PME2, pectin methylesterase 2 |
-0.164 |
-0.064 |
| 260 |
255710_at |
AT4G00030
|
[Plastid-lipid associated protein PAP / fibrillin family protein] |
-0.164 |
-0.016 |
| 261 |
256887_at |
AT3G15150
|
ATMMS21, A. THALIANA METHYL METHANE SULFONATE SENSITIVITY 21, HPY2, HIGH PLOIDY2, MMS21, METHYL METHANE SULFONATE SENSITIVITY 2 |
-0.164 |
-0.024 |
| 262 |
262973_at |
AT1G75600
|
[Histone superfamily protein] |
-0.163 |
0.022 |
| 263 |
266020_at |
AT2G05900
|
SDG11, SET domain protein 11, SUVH10, SU(VAR)3-9 HOMOLOG 10 |
-0.163 |
-0.002 |