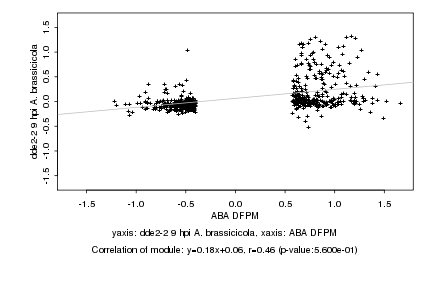
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ABA DFPM |
Log2 signal ratio
dde2-2 9 hpi A. brassicicola |
| 1 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
1.659 |
-0.025 |
| 2 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.514 |
0.002 |
| 3 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
1.487 |
-0.342 |
| 4 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
1.428 |
0.558 |
| 5 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
1.423 |
0.024 |
| 6 |
267559_at |
AT2G45570
|
CYP76C2, cytochrome P450, family 76, subfamily C, polypeptide 2 |
1.402 |
0.304 |
| 7 |
266544_at |
AT2G35300
|
AtLEA4-2, late embryogenesis abundant 4-2, LEA18, late embryogenesis abundant 18, LEA4-2, late embryogenesis abundant 4-2 |
1.379 |
-0.036 |
| 8 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
1.379 |
0.078 |
| 9 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
1.351 |
-0.210 |
| 10 |
267026_at |
AT2G38340
|
DREB19, dehydration response element-binding protein 19 |
1.335 |
0.605 |
| 11 |
258347_at |
AT3G17520
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.308 |
0.024 |
| 12 |
251456_at |
AT3G60120
|
BGLU27, beta glucosidase 27 |
1.299 |
0.464 |
| 13 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
1.288 |
0.080 |
| 14 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
1.286 |
0.004 |
| 15 |
251971_at |
AT3G53160
|
UGT73C7, UDP-glucosyl transferase 73C7 |
1.284 |
0.075 |
| 16 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
1.277 |
0.109 |
| 17 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
1.262 |
1.038 |
| 18 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
1.252 |
-0.144 |
| 19 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
1.240 |
-0.059 |
| 20 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
1.229 |
0.910 |
| 21 |
248227_at |
AT5G53820
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.212 |
0.020 |
| 22 |
262679_at |
AT1G75830
|
LCR67, low-molecular-weight cysteine-rich 67, PDF1.1, PLANT DEFENSIN 1.1 |
1.211 |
0.337 |
| 23 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
1.209 |
0.092 |
| 24 |
247061_at |
AT5G66780
|
unknown |
1.209 |
-0.023 |
| 25 |
260706_at |
AT1G32350
|
AOX1D, alternative oxidase 1D |
1.201 |
1.284 |
| 26 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
1.199 |
0.182 |
| 27 |
258224_at |
AT3G15670
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.195 |
-0.050 |
| 28 |
254189_at |
AT4G24000
|
ATCSLG2, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G2, CSLG2, cellulose synthase like G2 |
1.194 |
0.021 |
| 29 |
261243_at |
AT1G20180
|
unknown |
1.194 |
0.015 |
| 30 |
254413_at |
AT4G21440
|
ATM4, A. THALIANA MYB 4, ATMYB102, MYB-like 102, MYB102, MYB102, MYB-like 102 |
1.187 |
-0.024 |
| 31 |
266462_at |
AT2G47770
|
ATTSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related, TSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related |
1.169 |
0.097 |
| 32 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
1.163 |
1.320 |
| 33 |
258325_at |
AT3G22830
|
AT-HSFA6B, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A6B, HSFA6B, heat shock transcription factor A6B |
1.163 |
0.003 |
| 34 |
265024_at |
AT1G24600
|
unknown |
1.158 |
0.320 |
| 35 |
251824_at |
AT3G55090
|
ABCG16, ATP-binding cassette G16 |
1.153 |
0.022 |
| 36 |
257846_at |
AT3G12910
|
[NAC (No Apical Meristem) domain transcriptional regulator superfamily protein] |
1.147 |
0.785 |
| 37 |
256833_at |
AT3G22910
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
1.116 |
0.151 |
| 38 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
1.113 |
0.380 |
| 39 |
249333_at |
AT5G40990
|
GLIP1, GDSL lipase 1 |
1.113 |
1.310 |
| 40 |
264433_at |
AT1G61810
|
BGLU45, beta-glucosidase 45 |
1.090 |
0.510 |
| 41 |
260394_at |
AT1G74080
|
ATMYB122, MYB DOMAIN PROTEIN 122, MYB122, myb domain protein 122 |
1.088 |
0.623 |
| 42 |
262448_at |
AT1G49450
|
[Transducin/WD40 repeat-like superfamily protein] |
1.084 |
0.015 |
| 43 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
1.083 |
0.169 |
| 44 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
1.079 |
1.121 |
| 45 |
257540_at |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
1.072 |
0.967 |
| 46 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
1.067 |
0.642 |
| 47 |
265482_at |
AT2G15780
|
[Cupredoxin superfamily protein] |
1.060 |
0.101 |
| 48 |
267080_at |
AT2G41190
|
[Transmembrane amino acid transporter family protein] |
1.060 |
-0.057 |
| 49 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
1.059 |
0.160 |
| 50 |
266274_at |
AT2G29380
|
HAI3, highly ABA-induced PP2C gene 3 |
1.051 |
0.001 |
| 51 |
265723_at |
AT2G32140
|
[transmembrane receptors] |
1.044 |
0.739 |
| 52 |
261006_at |
AT1G26410
|
[FAD-binding Berberine family protein] |
1.037 |
1.104 |
| 53 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
1.032 |
-0.047 |
| 54 |
265216_at |
AT1G05100
|
MAPKKK18, mitogen-activated protein kinase kinase kinase 18 |
1.030 |
0.078 |
| 55 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
1.029 |
0.579 |
| 56 |
249848_at |
AT5G23220
|
NIC3, nicotinamidase 3 |
1.029 |
-0.010 |
| 57 |
250415_at |
AT5G11210
|
ATGLR2.5, ARABIDOPSIS THALIANA GLU, GLR2.5, glutamate receptor 2.5 |
1.023 |
0.499 |
| 58 |
252004_at |
AT3G52780
|
ATPAP20, PAP20 |
1.022 |
-0.013 |
| 59 |
255250_at |
AT4G05100
|
AtMYB74, myb domain protein 74, MYB74, myb domain protein 74 |
1.022 |
-0.054 |
| 60 |
266900_at |
AT2G34610
|
unknown |
1.004 |
-0.041 |
| 61 |
246406_at |
AT1G57650
|
[ATP binding] |
1.001 |
0.345 |
| 62 |
256296_at |
AT1G69480
|
[EXS (ERD1/XPR1/SYG1) family protein] |
0.998 |
-0.096 |
| 63 |
263385_at |
AT2G40170
|
ATEM6, ARABIDOPSIS EARLY METHIONINE-LABELLED 6, EM6, EARLY METHIONINE-LABELLED 6, GEA6, LATE EMBRYOGENESIS ABUNDANT 6 |
0.990 |
0.056 |
| 64 |
265084_at |
AT1G03790
|
AtTZF4, SOM, SOMNUS, TZF4, Tandem CCCH Zinc Finger protein 4 |
0.986 |
-0.067 |
| 65 |
261763_at |
AT1G15520
|
ABCG40, ATP-binding cassette G40, ATABCG40, Arabidopsis thaliana ATP-binding cassette G40, ATPDR12, PLEIOTROPIC DRUG RESISTANCE 12, PDR12, pleiotropic drug resistance 12 |
0.984 |
0.799 |
| 66 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.984 |
0.022 |
| 67 |
265658_at |
AT2G13810
|
ALD1, AGD2-like defense response protein 1, AtALD1, EDTS5, eds two suppressor 5 |
0.983 |
0.522 |
| 68 |
251743_at |
AT3G55890
|
[Yippee family putative zinc-binding protein] |
0.981 |
0.080 |
| 69 |
267263_at |
AT2G23110
|
[Late embryogenesis abundant protein, group 6] |
0.974 |
-0.030 |
| 70 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.966 |
0.742 |
| 71 |
259418_at |
AT1G02390
|
ATGPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2, GPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2 |
0.965 |
0.303 |
| 72 |
252487_at |
AT3G46660
|
UGT76E12, UDP-glucosyl transferase 76E12 |
0.964 |
0.018 |
| 73 |
251791_at |
AT3G55500
|
ATEXP16, ATEXPA16, expansin A16, ATHEXP ALPHA 1.7, EXP16, EXPANSIN 16, EXPA16, expansin A16 |
0.964 |
-0.116 |
| 74 |
256319_at |
AT1G35910
|
TPPD, trehalose-6-phosphate phosphatase D |
0.963 |
0.021 |
| 75 |
248959_at |
AT5G45630
|
unknown |
0.954 |
0.012 |
| 76 |
245982_at |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
0.949 |
-0.101 |
| 77 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.947 |
0.894 |
| 78 |
252320_at |
AT3G48580
|
XTH11, xyloglucan endotransglucosylase/hydrolase 11 |
0.947 |
0.013 |
| 79 |
251970_at |
AT3G53150
|
UGT73D1, UDP-glucosyl transferase 73D1 |
0.945 |
0.578 |
| 80 |
260357_at |
AT1G69260
|
AFP1, ABI five binding protein |
0.939 |
0.016 |
| 81 |
249896_at |
AT5G22530
|
unknown |
0.928 |
0.652 |
| 82 |
251766_at |
AT3G55910
|
unknown |
0.926 |
-0.058 |
| 83 |
258203_at |
AT3G13950
|
unknown |
0.924 |
0.946 |
| 84 |
249495_at |
AT5G39100
|
GLP6, germin-like protein 6 |
0.923 |
0.103 |
| 85 |
260406_at |
AT1G69920
|
ATGSTU12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 12, GSTU12, glutathione S-transferase TAU 12 |
0.916 |
0.631 |
| 86 |
248352_at |
AT5G52300
|
LTI65, LOW-TEMPERATURE-INDUCED 65, RD29B, RESPONSIVE TO DESSICATION 29B |
0.913 |
-0.054 |
| 87 |
266690_at |
AT2G19900
|
ATNADP-ME1, Arabidopsis thaliana NADP-malic enzyme 1, NADP-ME1, NADP-malic enzyme 1 |
0.913 |
-0.071 |
| 88 |
259439_at |
AT1G01480
|
ACS2, 1-amino-cyclopropane-1-carboxylate synthase 2, AT-ACC2 |
0.912 |
0.683 |
| 89 |
265796_at |
AT2G35730
|
[Heavy metal transport/detoxification superfamily protein ] |
0.909 |
0.183 |
| 90 |
254562_at |
AT4G19230
|
CYP707A1, cytochrome P450, family 707, subfamily A, polypeptide 1 |
0.909 |
-0.058 |
| 91 |
250624_at |
AT5G07330
|
unknown |
0.908 |
-0.015 |
| 92 |
253796_at |
AT4G28460
|
unknown |
0.907 |
1.169 |
| 93 |
249001_at |
AT5G44990
|
[Glutathione S-transferase family protein] |
0.907 |
0.602 |
| 94 |
252006_at |
AT3G52820
|
ATPAP22, PURPLE ACID PHOSPHATASE 22, PAP22, purple acid phosphatase 22 |
0.900 |
-0.036 |
| 95 |
251293_at |
AT3G61930
|
unknown |
0.898 |
0.217 |
| 96 |
252321_at |
AT3G48510
|
unknown |
0.895 |
-0.000 |
| 97 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
0.893 |
0.352 |
| 98 |
248209_at |
AT5G53990
|
[UDP-Glycosyltransferase superfamily protein] |
0.881 |
-0.002 |
| 99 |
259705_at |
AT1G77450
|
anac032, NAC domain containing protein 32, NAC032, NAC domain containing protein 32 |
0.880 |
0.369 |
| 100 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
0.878 |
-0.096 |
| 101 |
248896_at |
AT5G46350
|
ATWRKY8, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 8, WRKY8, WRKY DNA-binding protein 8 |
0.877 |
0.456 |
| 102 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.875 |
0.277 |
| 103 |
259975_at |
AT1G76470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.872 |
0.111 |
| 104 |
249770_at |
AT5G24110
|
ATWRKY30, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 30, WRKY30, WRKY DNA-binding protein 30 |
0.872 |
1.061 |
| 105 |
250230_at |
AT5G13900
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.870 |
-0.081 |
| 106 |
254597_at |
AT4G18980
|
AtS40-3, AtS40-3 |
0.869 |
0.015 |
| 107 |
247957_at |
AT5G57050
|
ABI2, ABA INSENSITIVE 2, AtABI2 |
0.869 |
0.157 |
| 108 |
245363_at |
AT4G15120
|
[VQ motif-containing protein] |
0.868 |
0.560 |
| 109 |
246988_at |
AT5G67340
|
[ARM repeat superfamily protein] |
0.862 |
0.584 |
| 110 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.858 |
0.458 |
| 111 |
245748_at |
AT1G51140
|
AKS1, ABA-responsive kinase substrate 1, FBH3, FLOWERING BHLH 3 |
0.858 |
-0.302 |
| 112 |
261249_at |
AT1G05880
|
ARI12, ARIADNE 12, ATARI12, ARABIDOPSIS ARIADNE 12 |
0.857 |
0.504 |
| 113 |
245738_at |
AT1G44130
|
[Eukaryotic aspartyl protease family protein] |
0.857 |
0.759 |
| 114 |
253044_at |
AT4G37290
|
unknown |
0.854 |
1.233 |
| 115 |
250541_at |
AT5G09520
|
PELPK2, Pro-Glu-Leu|Ile|Val-Pro-Lys 2 |
0.854 |
0.034 |
| 116 |
248700_at |
AT5G48400
|
ATGLR1.2, GLR1.2, GLUTAMATE RECEPTOR 1.2 |
0.845 |
0.573 |
| 117 |
254905_at |
AT4G11170
|
RMG1, Resistance Methylated Gene 1 |
0.842 |
0.620 |
| 118 |
259987_at |
AT1G75030
|
ATLP-3, thaumatin-like protein 3, TLP-3, thaumatin-like protein 3 |
0.840 |
-0.080 |
| 119 |
252482_at |
AT3G46670
|
UGT76E11, UDP-glucosyl transferase 76E11 |
0.840 |
-0.050 |
| 120 |
260803_at |
AT1G78340
|
ATGSTU22, glutathione S-transferase TAU 22, GSTU22, glutathione S-transferase TAU 22 |
0.839 |
0.110 |
| 121 |
252073_at |
AT3G51750
|
unknown |
0.834 |
-0.102 |
| 122 |
259150_at |
AT3G10320
|
[Glycosyltransferase family 61 protein] |
0.832 |
0.102 |
| 123 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
0.825 |
-0.097 |
| 124 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
0.824 |
0.022 |
| 125 |
263363_at |
AT2G03850
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.824 |
0.025 |
| 126 |
260561_at |
AT2G43580
|
[Chitinase family protein] |
0.824 |
0.125 |
| 127 |
256176_at |
AT1G51640
|
ATEXO70G2, exocyst subunit exo70 family protein G2, EXO70G2, exocyst subunit exo70 family protein G2 |
0.823 |
-0.096 |
| 128 |
258652_at |
AT3G09910
|
ATRAB18C, ATRABC2B, RAB GTPase homolog C2B, RABC2b, RAB GTPase homolog C2B |
0.823 |
-0.006 |
| 129 |
246183_at |
AT5G20940
|
[Glycosyl hydrolase family protein] |
0.823 |
0.022 |
| 130 |
256285_at |
AT3G12510
|
[MADS-box family protein] |
0.821 |
0.194 |
| 131 |
256114_at |
AT1G16850
|
unknown |
0.821 |
-0.163 |
| 132 |
246984_at |
AT5G67310
|
CYP81G1, cytochrome P450, family 81, subfamily G, polypeptide 1 |
0.815 |
0.752 |
| 133 |
265983_at |
AT2G18550
|
ATHB21, homeobox protein 21, HB-2, homeobox-2, HB21, homeobox protein 21 |
0.814 |
0.001 |
| 134 |
251028_at |
AT5G02230
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.812 |
0.036 |
| 135 |
251668_at |
AT3G57010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.812 |
-0.026 |
| 136 |
252209_at |
AT3G50400
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.809 |
-0.085 |
| 137 |
254598_at |
AT4G18990
|
XTH29, xyloglucan endotransglucosylase/hydrolase 29 |
0.807 |
0.466 |
| 138 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.807 |
-0.073 |
| 139 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
0.807 |
0.049 |
| 140 |
264318_at |
AT1G04220
|
KCS2, 3-ketoacyl-CoA synthase 2 |
0.804 |
-0.043 |
| 141 |
245734_at |
AT1G73480
|
[alpha/beta-Hydrolases superfamily protein] |
0.804 |
0.134 |
| 142 |
261449_at |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
0.804 |
1.307 |
| 143 |
265501_at |
AT2G15490
|
UGT73B4, UDP-glycosyltransferase 73B4 |
0.799 |
0.481 |
| 144 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.796 |
0.020 |
| 145 |
252345_at |
AT3G48640
|
unknown |
0.793 |
0.541 |
| 146 |
248545_at |
AT5G50260
|
CEP1, cysteine endopeptidase 1 |
0.792 |
0.013 |
| 147 |
252334_at |
AT3G48850
|
MPT2, mitochondrial phosphate transporter 2, PHT3;2, phosphate transporter 3;2 |
0.792 |
0.806 |
| 148 |
266269_at |
AT2G29480
|
ATGSTU2, glutathione S-transferase tau 2, GST20, GLUTATHIONE S-TRANSFERASE 20, GSTU2, glutathione S-transferase tau 2 |
0.790 |
0.048 |
| 149 |
265913_at |
AT2G25625
|
unknown |
0.786 |
-0.054 |
| 150 |
266995_at |
AT2G34500
|
CYP710A1, cytochrome P450, family 710, subfamily A, polypeptide 1 |
0.783 |
1.004 |
| 151 |
260163_at |
AT1G79900
|
ATMBAC2, RABIDOPSIS MITOCHONDRIAL BASIC AMINO ACID CARRIER 2, BAC2 |
0.783 |
-0.058 |
| 152 |
262454_at |
AT1G11190
|
BFN1, bifunctional nuclease i, ENDO1, ENDONUCLEASE 1 |
0.783 |
-0.018 |
| 153 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.783 |
0.869 |
| 154 |
249860_at |
AT5G22860
|
[Serine carboxypeptidase S28 family protein] |
0.782 |
0.042 |
| 155 |
251334_at |
AT3G61390
|
[RING/U-box superfamily protein] |
0.781 |
0.012 |
| 156 |
264217_at |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
0.781 |
-0.043 |
| 157 |
248505_at |
AT5G50360
|
unknown |
0.777 |
-0.035 |
| 158 |
255521_at |
AT4G02280
|
ATSUS3, SUS3, sucrose synthase 3 |
0.774 |
-0.033 |
| 159 |
245082_at |
AT2G23270
|
unknown |
0.771 |
0.980 |
| 160 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
0.770 |
0.022 |
| 161 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.755 |
1.270 |
| 162 |
260040_at |
AT1G68765
|
IDA, INFLORESCENCE DEFICIENT IN ABSCISSION |
0.754 |
0.933 |
| 163 |
259033_at |
AT3G09405
|
[Pectinacetylesterase family protein] |
0.752 |
0.726 |
| 164 |
266052_at |
AT2G40740
|
ATWRKY55, WRKY DNA-BINDING PROTEIN 55, WRKY55, WRKY DNA-binding protein 55 |
0.751 |
0.655 |
| 165 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
0.748 |
-0.082 |
| 166 |
250239_at |
AT5G13580
|
ABCG6, ATP-binding cassette G6 |
0.748 |
-0.018 |
| 167 |
251096_at |
AT5G01550
|
LecRK-VI.3, L-type lectin receptor kinase VI.3, LECRKA4.2, lectin receptor kinase a4.1 |
0.746 |
0.741 |
| 168 |
265119_at |
AT1G62570
|
FMO GS-OX4, flavin-monooxygenase glucosinolate S-oxygenase 4 |
0.746 |
-0.045 |
| 169 |
266078_at |
AT2G40670
|
ARR16, response regulator 16, RR16, response regulator 16 |
0.744 |
-0.070 |
| 170 |
262347_at |
AT1G64110
|
DAA1, DUO1-activated ATPase 1 |
0.743 |
0.010 |
| 171 |
263005_at |
AT1G54540
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.742 |
-0.021 |
| 172 |
255074_at |
AT4G09100
|
[RING/U-box superfamily protein] |
0.742 |
0.014 |
| 173 |
261240_at |
AT1G32940
|
ATSBT3.5, SBT3.5 |
0.742 |
0.661 |
| 174 |
262640_at |
AT1G62760
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.737 |
-0.048 |
| 175 |
250474_at |
AT5G10230
|
ANN7, ANNEXIN 7, ANNAT7, annexin 7 |
0.737 |
0.033 |
| 176 |
261899_at |
AT1G80820
|
ATCCR2, CCR2, cinnamoyl coa reductase |
0.736 |
0.770 |
| 177 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.733 |
1.180 |
| 178 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.732 |
0.032 |
| 179 |
262164_at |
AT1G78070
|
[Transducin/WD40 repeat-like superfamily protein] |
0.731 |
-0.030 |
| 180 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.730 |
-0.030 |
| 181 |
256378_at |
AT1G66830
|
[Leucine-rich repeat protein kinase family protein] |
0.730 |
0.093 |
| 182 |
248333_at |
AT5G52390
|
[PAR1 protein] |
0.728 |
-0.515 |
| 183 |
246241_at |
AT4G37050
|
AtPLAIVC, phospholipase A IVC, PLA V, PLAIII{beta}, patatin-related phospholipase III beta, PLP4, PATATIN-like protein 4 |
0.726 |
-0.069 |
| 184 |
252400_at |
AT3G48020
|
unknown |
0.725 |
0.471 |
| 185 |
249771_at |
AT5G24080
|
[Protein kinase superfamily protein] |
0.721 |
-0.077 |
| 186 |
245349_at |
AT4G16690
|
ATMES16, ARABIDOPSIS THALIANA METHYL ESTERASE 16, MES16, methyl esterase 16 |
0.721 |
-0.298 |
| 187 |
254575_at |
AT4G19460
|
[UDP-Glycosyltransferase superfamily protein] |
0.720 |
0.116 |
| 188 |
260881_at |
AT1G21550
|
[Calcium-binding EF-hand family protein] |
0.716 |
0.124 |
| 189 |
252421_at |
AT3G47540
|
[Chitinase family protein] |
0.713 |
0.201 |
| 190 |
263881_at |
AT2G21820
|
unknown |
0.712 |
-0.019 |
| 191 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
0.712 |
0.851 |
| 192 |
253851_at |
AT4G28110
|
AtMYB41, myb domain protein 41, MYB41, myb domain protein 41 |
0.712 |
-0.014 |
| 193 |
252019_at |
AT3G53040
|
[late embryogenesis abundant protein, putative / LEA protein, putative] |
0.711 |
0.051 |
| 194 |
259618_at |
AT1G48000
|
AtMYB112, myb domain protein 112, MYB112, myb domain protein 112 |
0.710 |
-0.009 |
| 195 |
259428_at |
AT1G01560
|
ATMPK11, MAP kinase 11, MPK11, MAP kinase 11 |
0.708 |
0.513 |
| 196 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
0.705 |
-0.401 |
| 197 |
253152_at |
AT4G35690
|
unknown |
0.705 |
0.036 |
| 198 |
262244_at |
AT1G48260
|
CIPK17, CBL-interacting protein kinase 17, SnRK3.21, SNF1-RELATED PROTEIN KINASE 3.21 |
0.704 |
-0.074 |
| 199 |
254014_at |
AT4G26120
|
[Ankyrin repeat family protein / BTB/POZ domain-containing protein] |
0.704 |
0.257 |
| 200 |
260841_at |
AT1G29195
|
unknown |
0.702 |
0.023 |
| 201 |
262942_at |
AT1G79450
|
ALIS5, ALA-interacting subunit 5 |
0.697 |
0.338 |
| 202 |
266170_at |
AT2G39050
|
ArathEULS3, EULS3, Euonymus lectin S3 |
0.696 |
-0.096 |
| 203 |
261366_at |
AT1G53100
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
0.696 |
-0.024 |
| 204 |
259442_at |
AT1G02310
|
MAN1, endo-beta-mannanase 1 |
0.695 |
0.004 |
| 205 |
265618_at |
AT2G25460
|
unknown |
0.694 |
0.098 |
| 206 |
257271_at |
AT3G28007
|
AtSWEET4, SWEET4 |
0.693 |
-0.010 |
| 207 |
249881_at |
AT5G23190
|
CYP86B1, cytochrome P450, family 86, subfamily B, polypeptide 1 |
0.693 |
0.021 |
| 208 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.687 |
-0.011 |
| 209 |
251156_at |
AT3G63060
|
EDL3, EID1-like 3 |
0.685 |
-0.017 |
| 210 |
255881_at |
AT1G67070
|
DIN9, DARK INDUCIBLE 9, PMI2, PHOSPHOMANNOSE ISOMERASE 2 |
0.685 |
0.070 |
| 211 |
259231_at |
AT3G11410
|
AHG3, ATPP2CA, ARABIDOPSIS THALIANA PROTEIN PHOSPHATASE 2CA, PP2CA, protein phosphatase 2CA |
0.684 |
-0.023 |
| 212 |
247215_at |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
0.684 |
1.170 |
| 213 |
255723_at |
AT3G29575
|
AFP3, ABI five binding protein 3 |
0.682 |
-0.127 |
| 214 |
260845_at |
AT1G17310
|
[MADS-box transcription factor family protein] |
0.680 |
-0.046 |
| 215 |
248424_at |
AT5G51680
|
[hydroxyproline-rich glycoprotein family protein] |
0.680 |
0.023 |
| 216 |
251545_at |
AT3G58810
|
ATMTP3, ARABIDOPSIS METAL TOLERANCE PROTEIN 3, ATMTPA2, MTP3, METAL TOLERANCE PROTEIN 3, MTPA2, metal tolerance protein A2 |
0.676 |
-0.015 |
| 217 |
246582_at |
AT1G31750
|
[proline-rich family protein] |
0.675 |
-0.057 |
| 218 |
254234_at |
AT4G23680
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.674 |
-0.040 |
| 219 |
246370_at |
AT1G51920
|
unknown |
0.673 |
1.102 |
| 220 |
267372_at |
AT2G26290
|
ARSK1, root-specific kinase 1 |
0.672 |
0.078 |
| 221 |
254289_at |
AT4G22980
|
unknown |
0.669 |
0.212 |
| 222 |
266780_at |
AT2G29110
|
ATGLR2.8, glutamate receptor 2.8, GLR2.8, GLR2.8, glutamate receptor 2.8 |
0.668 |
0.123 |
| 223 |
264580_at |
AT1G05340
|
unknown |
0.667 |
1.103 |
| 224 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.665 |
0.946 |
| 225 |
263825_at |
AT2G40370
|
LAC5, laccase 5 |
0.663 |
-0.040 |
| 226 |
249889_at |
AT5G22540
|
unknown |
0.661 |
0.278 |
| 227 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
0.661 |
1.185 |
| 228 |
260386_at |
AT1G74010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.660 |
0.208 |
| 229 |
260239_at |
AT1G74360
|
[Leucine-rich repeat protein kinase family protein] |
0.658 |
0.775 |
| 230 |
262085_at |
AT1G56060
|
unknown |
0.657 |
0.750 |
| 231 |
259316_at |
AT3G01175
|
unknown |
0.657 |
0.029 |
| 232 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.657 |
0.972 |
| 233 |
253494_at |
AT4G31830
|
unknown |
0.653 |
0.028 |
| 234 |
262580_at |
AT1G15330
|
[Cystathionine beta-synthase (CBS) protein] |
0.650 |
-0.090 |
| 235 |
259743_at |
AT1G71140
|
[MATE efflux family protein] |
0.650 |
0.015 |
| 236 |
267391_at |
AT2G44480
|
BGLU17, beta glucosidase 17 |
0.649 |
0.050 |
| 237 |
250158_at |
AT5G15190
|
unknown |
0.648 |
0.086 |
| 238 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
0.645 |
1.157 |
| 239 |
260261_at |
AT1G68450
|
PDE337, PIGMENT DEFECTIVE 337 |
0.643 |
-0.000 |
| 240 |
259149_at |
AT3G10340
|
PAL4, phenylalanine ammonia-lyase 4 |
0.636 |
0.162 |
| 241 |
250279_at |
AT5G13200
|
[GRAM domain family protein] |
0.636 |
0.309 |
| 242 |
259282_at |
AT3G11430
|
ATGPAT5, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 5, GPAT5, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 5 |
0.635 |
0.072 |
| 243 |
248701_at |
AT5G48410
|
ATGLR1.3, ARABIDOPSIS THALIANA GLUTAMATE RECEPTOR 1.3, GLR1.3, glutamate receptor 1.3 |
0.635 |
0.386 |
| 244 |
245412_at |
AT4G17280
|
[Auxin-responsive family protein] |
0.635 |
-0.034 |
| 245 |
266503_at |
AT2G47780
|
[Rubber elongation factor protein (REF)] |
0.633 |
-0.021 |
| 246 |
267140_at |
AT2G38250
|
[Homeodomain-like superfamily protein] |
0.633 |
0.162 |
| 247 |
248154_at |
AT5G54400
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.633 |
-0.101 |
| 248 |
257978_at |
AT3G20860
|
ATNEK5, NIMA-related kinase 5, NEK5, NIMA-related kinase 5 |
0.632 |
0.478 |
| 249 |
260581_at |
AT2G47190
|
ATMYB2, MYB DOMAIN PROTEIN 2, MYB2, myb domain protein 2 |
0.630 |
0.116 |
| 250 |
267058_at |
AT2G32510
|
MAPKKK17, mitogen-activated protein kinase kinase kinase 17 |
0.629 |
-0.012 |
| 251 |
247312_at |
AT5G63970
|
RGLG3, RING DOMAIN LIGASE 3 |
0.629 |
-0.075 |
| 252 |
250576_at |
AT5G08250
|
[Cytochrome P450 superfamily protein] |
0.628 |
-0.025 |
| 253 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
0.626 |
-0.314 |
| 254 |
251207_at |
AT3G63050
|
unknown |
0.623 |
-0.039 |
| 255 |
255575_at |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
0.623 |
-0.010 |
| 256 |
245904_at |
AT5G11110
|
ATSPS2F, sucrose phosphate synthase 2F, KNS2, KAONASHI 2, SPS1, SUCROSE PHOSPHATE SYNTHASE 1, SPS2F, sucrose phosphate synthase 2F, SPSA2, sucrose-phosphate synthase A2 |
0.623 |
0.033 |
| 257 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
0.620 |
0.045 |
| 258 |
245662_at |
AT1G28190
|
unknown |
0.619 |
0.737 |
| 259 |
248230_at |
AT5G53830
|
[VQ motif-containing protein] |
0.619 |
0.209 |
| 260 |
252137_at |
AT3G50980
|
XERO1, dehydrin xero 1 |
0.618 |
-0.006 |
| 261 |
254710_at |
AT4G18050
|
ABCB9, ATP-binding cassette B9, PGP9, P-glycoprotein 9 |
0.617 |
-0.019 |
| 262 |
264891_at |
AT1G23200
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.616 |
0.162 |
| 263 |
251137_at |
AT5G01300
|
[PEBP (phosphatidylethanolamine-binding protein) family protein] |
0.616 |
0.023 |
| 264 |
264288_at |
AT1G62045
|
unknown |
0.615 |
0.052 |
| 265 |
259922_at |
AT1G72770
|
AtHAB1, HAB1, HYPERSENSITIVE TO ABA1 |
0.614 |
-0.129 |
| 266 |
259297_at |
AT3G05360
|
AtRLP30, receptor like protein 30, RLP30, receptor like protein 30 |
0.613 |
0.527 |
| 267 |
248916_at |
AT5G45840
|
[Leucine-rich repeat protein kinase family protein] |
0.613 |
0.133 |
| 268 |
248553_at |
AT5G50170
|
[C2 calcium/lipid-binding and GRAM domain containing protein] |
0.612 |
-0.145 |
| 269 |
247618_at |
AT5G60280
|
LecRK-I.8, L-type lectin receptor kinase I.8 |
0.608 |
0.413 |
| 270 |
267496_at |
AT2G30550
|
DALL3, DAD1-Like Lipase 3 |
0.607 |
0.282 |
| 271 |
254201_at |
AT4G24130
|
unknown |
0.606 |
0.000 |
| 272 |
265672_at |
AT2G31980
|
AtCYS2, PHYTOCYSTATIN 2, CYS2, PHYTOCYSTATIN 2 |
0.604 |
-0.044 |
| 273 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.604 |
0.863 |
| 274 |
256877_at |
AT3G26470
|
[Powdery mildew resistance protein, RPW8 domain] |
0.600 |
0.198 |
| 275 |
248207_at |
AT5G53970
|
TAT7, tyrosine aminotransferase 7 |
0.599 |
0.114 |
| 276 |
253963_at |
AT4G26470
|
[Calcium-binding EF-hand family protein] |
0.598 |
0.093 |
| 277 |
253293_at |
AT4G33905
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
0.597 |
-0.004 |
| 278 |
246485_at |
AT5G16080
|
AtCXE17, carboxyesterase 17, CXE17, carboxyesterase 17 |
0.597 |
0.140 |
| 279 |
265499_at |
AT2G15480
|
UGT73B5, UDP-glucosyl transferase 73B5 |
0.597 |
0.727 |
| 280 |
256354_at |
AT1G54870
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.595 |
-0.072 |
| 281 |
248248_at |
AT5G53120
|
ATSPDS3, SPERMIDINE SYNTHASE 3, SPDS3, spermidine synthase 3, SPMS |
0.595 |
-0.013 |
| 282 |
264436_at |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
0.594 |
0.105 |
| 283 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
0.594 |
0.321 |
| 284 |
266346_at |
AT2G01430
|
ATHB-17, ARABIDOPSIS THALIANA HOMEOBOX-LEUCINE ZIPPER PROTEIN 17, ATHB17, homeobox-leucine zipper protein 17, HB17, homeobox-leucine zipper protein 17 |
0.592 |
-0.062 |
| 285 |
261618_at |
AT1G33110
|
[MATE efflux family protein] |
0.592 |
0.326 |
| 286 |
251944_at |
AT3G53510
|
ABCG20, ATP-binding cassette G20 |
0.589 |
-0.061 |
| 287 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.586 |
0.020 |
| 288 |
260039_at |
AT1G68795
|
CLE12, CLAVATA3/ESR-RELATED 12 |
0.586 |
-0.002 |
| 289 |
254062_at |
AT4G25380
|
AtSAP10, Arabidopsis thaliana stress-associated protein 10, SAP10, stress-associated protein 10 |
0.583 |
0.173 |
| 290 |
249767_at |
AT5G24090
|
ATCHIA, chitinase A, CHIA, chitinase A |
0.580 |
0.264 |
| 291 |
260648_at |
AT1G08050
|
[Zinc finger (C3HC4-type RING finger) family protein] |
0.579 |
0.444 |
| 292 |
248761_at |
AT5G47635
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.579 |
0.012 |
| 293 |
265728_at |
AT2G31990
|
[Exostosin family protein] |
0.579 |
0.277 |
| 294 |
256757_at |
AT3G25620
|
ABCG21, ATP-binding cassette G21 |
0.579 |
-0.004 |
| 295 |
260415_at |
AT1G69790
|
[Protein kinase superfamily protein] |
0.578 |
0.419 |
| 296 |
256073_at |
AT1G18100
|
E12A11, MFT, MOTHER OF FT AND TFL1 |
0.575 |
0.038 |
| 297 |
253710_at |
AT4G29230
|
anac075, NAC domain containing protein 75, NAC075, NAC domain containing protein 75 |
0.575 |
0.009 |
| 298 |
257466_at |
AT1G62840
|
unknown |
0.573 |
0.140 |
| 299 |
247706_at |
AT5G59480
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.571 |
-0.223 |
| 300 |
250165_at |
AT5G15290
|
CASP5, Casparian strip membrane domain protein 5 |
0.569 |
0.001 |
| 301 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-1.221 |
0.008 |
| 302 |
246004_at |
AT5G20630
|
ATGER3, ARABIDOPSIS THALIANA GERMIN 3, GER3, germin 3, GLP3, GERMIN-LIKE PROTEIN 3, GLP3A, GLP3B |
-1.201 |
-0.062 |
| 303 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
-1.110 |
-0.043 |
| 304 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
-1.083 |
-0.189 |
| 305 |
257021_at |
AT3G19710
|
BCAT4, branched-chain aminotransferase4 |
-1.070 |
-0.278 |
| 306 |
265400_at |
AT2G10940
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-1.069 |
-0.045 |
| 307 |
249866_at |
AT5G23010
|
IMS3, 2-ISOPROPYLMALATE SYNTHASE 3, MAM1, methylthioalkylmalate synthase 1 |
-1.043 |
-0.216 |
| 308 |
259813_at |
AT1G49860
|
ATGSTF14, glutathione S-transferase (class phi) 14, GSTF14, glutathione S-transferase (class phi) 14 |
-0.987 |
-0.030 |
| 309 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.971 |
0.109 |
| 310 |
254384_at |
AT4G21870
|
[HSP20-like chaperones superfamily protein] |
-0.958 |
-0.021 |
| 311 |
258055_at |
AT3G16250
|
NDF4, NDH-dependent cyclic electron flow 1, PnsB3, Photosynthetic NDH subcomplex B 3 |
-0.946 |
-0.105 |
| 312 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.913 |
-0.154 |
| 313 |
259839_at |
AT1G52190
|
AtNPF1.2, NPF1.2, NRT1/ PTR family 1.2, NRT1.11, nitrate transporter 1.11 |
-0.912 |
-0.012 |
| 314 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.911 |
0.188 |
| 315 |
267459_at |
AT2G33850
|
unknown |
-0.907 |
0.013 |
| 316 |
265342_at |
AT2G18300
|
HBI1, HOMOLOG OF BEE2 INTERACTING WITH IBH 1 |
-0.906 |
-0.123 |
| 317 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
-0.894 |
-0.035 |
| 318 |
262304_at |
AT1G70890
|
MLP43, MLP-like protein 43 |
-0.893 |
-0.139 |
| 319 |
255856_at |
AT1G66940
|
[protein kinase-related] |
-0.889 |
-0.004 |
| 320 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
-0.882 |
0.362 |
| 321 |
252183_at |
AT3G50740
|
UGT72E1, UDP-glucosyl transferase 72E1 |
-0.880 |
0.081 |
| 322 |
258935_at |
AT3G10120
|
unknown |
-0.856 |
-0.023 |
| 323 |
247362_at |
AT5G63140
|
ATPAP29, purple acid phosphatase 29, PAP29, PAP29, purple acid phosphatase 29 |
-0.832 |
-0.157 |
| 324 |
247377_at |
AT5G63180
|
[Pectin lyase-like superfamily protein] |
-0.820 |
-0.106 |
| 325 |
256796_at |
AT3G22210
|
unknown |
-0.820 |
-0.070 |
| 326 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
-0.815 |
-0.125 |
| 327 |
250006_at |
AT5G18660
|
PCB2, PALE-GREEN AND CHLOROPHYLL B REDUCED 2 |
-0.814 |
-0.143 |
| 328 |
263761_at |
AT2G21330
|
AtFBA1, FBA1, fructose-bisphosphate aldolase 1 |
-0.803 |
-0.039 |
| 329 |
259786_at |
AT1G29660
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.800 |
-0.109 |
| 330 |
252272_at |
AT3G49670
|
BAM2, BARELY ANY MERISTEM 2 |
-0.789 |
0.012 |
| 331 |
261981_at |
AT1G33811
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.787 |
0.008 |
| 332 |
246275_at |
AT4G36540
|
BEE2, BR enhanced expression 2 |
-0.775 |
-0.164 |
| 333 |
253971_at |
AT4G26530
|
AtFBA5, FBA5, fructose-bisphosphate aldolase 5 |
-0.770 |
-0.019 |
| 334 |
261480_at |
AT1G14280
|
PKS2, phytochrome kinase substrate 2 |
-0.767 |
-0.116 |
| 335 |
261927_at |
AT1G22500
|
AtATL15, Arabidopsis thaliana Arabidopsis toxicos en levadura 15, ATL15, Arabidopsis toxicos en levadura 15 |
-0.761 |
0.019 |
| 336 |
263034_at |
AT1G24020
|
MLP423, MLP-like protein 423 |
-0.748 |
-0.140 |
| 337 |
251935_at |
AT3G54090
|
FLN1, fructokinase-like 1 |
-0.747 |
-0.081 |
| 338 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
-0.745 |
-0.121 |
| 339 |
256736_at |
AT3G29410
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.736 |
0.017 |
| 340 |
246860_at |
AT5G25840
|
unknown |
-0.730 |
0.023 |
| 341 |
249008_at |
AT5G44680
|
[DNA glycosylase superfamily protein] |
-0.729 |
-0.057 |
| 342 |
251899_at |
AT3G54400
|
[Eukaryotic aspartyl protease family protein] |
-0.724 |
-0.021 |
| 343 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.724 |
0.185 |
| 344 |
254410_at |
AT4G21410
|
CRK29, cysteine-rich RLK (RECEPTOR-like protein kinase) 29 |
-0.723 |
0.364 |
| 345 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
-0.722 |
-0.104 |
| 346 |
257891_at |
AT3G17170
|
RFC3, REGULATOR OF FATTY-ACID COMPOSITION 3 |
-0.720 |
-0.102 |
| 347 |
245029_at |
AT2G26580
|
YAB5, YABBY5 |
-0.719 |
-0.090 |
| 348 |
264574_at |
AT1G05300
|
ZIP5, zinc transporter 5 precursor |
-0.714 |
0.262 |
| 349 |
258468_at |
AT3G06070
|
unknown |
-0.713 |
-0.077 |
| 350 |
254794_at |
AT4G12970
|
EPFL9, STOMAGEN, STOMAGEN |
-0.710 |
-0.096 |
| 351 |
247477_at |
AT5G62340
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.708 |
-0.031 |
| 352 |
261638_at |
AT1G49975
|
[INVOLVED IN: photosynthesis] |
-0.702 |
-0.099 |
| 353 |
246652_at |
AT5G35190
|
EXT13, extensin 13 |
-0.702 |
-0.052 |
| 354 |
247549_at |
AT5G61420
|
AtMYB28, HAG1, HIGH ALIPHATIC GLUCOSINOLATE 1, MYB28, myb domain protein 28, PMG1, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 1 |
-0.699 |
-0.113 |
| 355 |
262286_at |
AT1G68585
|
unknown |
-0.696 |
-0.074 |
| 356 |
248895_at |
AT5G46330
|
FLS2, FLAGELLIN-SENSITIVE 2 |
-0.696 |
0.258 |
| 357 |
248094_at |
AT5G55220
|
[trigger factor type chaperone family protein] |
-0.695 |
-0.156 |
| 358 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
-0.694 |
-0.222 |
| 359 |
263979_at |
AT2G42840
|
PDF1, protodermal factor 1 |
-0.693 |
0.023 |
| 360 |
253887_at |
AT4G27730
|
ATOPT6, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 6, OPT6, oligopeptide transporter 1 |
-0.692 |
0.178 |
| 361 |
260883_at |
AT1G29270
|
unknown |
-0.691 |
-0.035 |
| 362 |
247871_at |
AT5G57530
|
AtXTH12, XTH12, xyloglucan endotransglucosylase/hydrolase 12 |
-0.678 |
-0.059 |
| 363 |
252199_at |
AT3G50270
|
[HXXXD-type acyl-transferase family protein] |
-0.677 |
-0.035 |
| 364 |
264782_at |
AT1G08810
|
AtMYB60, myb domain protein 60, MYB60, myb domain protein 60 |
-0.677 |
-0.030 |
| 365 |
252853_at |
AT4G39710
|
FKBP16-2, FK506-binding protein 16-2, PnsL4, Photosynthetic NDH subcomplex L 4 |
-0.674 |
-0.075 |
| 366 |
252362_at |
AT3G48500
|
PDE312, PIGMENT DEFECTIVE 312, PTAC10, PLASTID TRANSCRIPTIONALLY ACTIVE 10, TAC10 |
-0.671 |
-0.178 |
| 367 |
255579_at |
AT4G01460
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.670 |
-0.185 |
| 368 |
262162_at |
AT1G78020
|
unknown |
-0.664 |
-0.061 |
| 369 |
253040_at |
AT4G37800
|
XTH7, xyloglucan endotransglucosylase/hydrolase 7 |
-0.662 |
-0.024 |
| 370 |
247755_at |
AT5G59090
|
ATSBT4.12, subtilase 4.12, SBT4.12, subtilase 4.12 |
-0.658 |
-0.107 |
| 371 |
255433_at |
AT4G03210
|
XTH9, xyloglucan endotransglucosylase/hydrolase 9 |
-0.654 |
-0.105 |
| 372 |
254687_at |
AT4G13770
|
CYP83A1, cytochrome P450, family 83, subfamily A, polypeptide 1, REF2, REDUCED EPIDERMAL FLUORESCENCE 2 |
-0.650 |
-0.157 |
| 373 |
248509_at |
AT5G50335
|
unknown |
-0.650 |
-0.075 |
| 374 |
266175_at |
AT2G02450
|
ANAC034, Arabidopsis NAC domain containing protein 34, ANAC035, NAC domain containing protein 35, AtLOV1, LOV1, LONG VEGETATIVE PHASE 1, NAC035, NAC domain containing protein 35 |
-0.646 |
-0.107 |
| 375 |
250801_at |
AT5G04960
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.645 |
0.021 |
| 376 |
266790_at |
AT2G28950
|
ATEXP6, ARABIDOPSIS THALIANA TEXPANSIN 6, ATEXPA6, expansin A6, ATHEXP ALPHA 1.8, EXPA6, expansin A6 |
-0.636 |
-0.085 |
| 377 |
251395_at |
AT2G45470
|
AGP8, ARABINOGALACTAN PROTEIN 8, FLA8, FASCICLIN-like arabinogalactan protein 8 |
-0.628 |
-0.073 |
| 378 |
260343_at |
AT1G69200
|
FLN2, fructokinase-like 2 |
-0.624 |
-0.178 |
| 379 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
-0.622 |
-0.144 |
| 380 |
256125_at |
AT1G18250
|
ATLP-1 |
-0.619 |
-0.158 |
| 381 |
247288_at |
AT5G64330
|
JK218, NPH3, NON-PHOTOTROPIC HYPOCOTYL 3, RPT3, ROOT PHOTOTROPISM 3 |
-0.619 |
0.024 |
| 382 |
259892_at |
AT1G72610
|
ATGER1, A. THALIANA GERMIN-LIKE PROTEIN 1, GER1, germin-like protein 1, GLP1, GERMIN-LIKE PROTEIN 1 |
-0.618 |
-0.019 |
| 383 |
255652_at |
AT4G00950
|
MEE47, maternal effect embryo arrest 47 |
-0.617 |
-0.119 |
| 384 |
254119_at |
AT4G24780
|
[Pectin lyase-like superfamily protein] |
-0.617 |
-0.057 |
| 385 |
250366_at |
AT5G11420
|
unknown |
-0.613 |
-0.027 |
| 386 |
263679_at |
AT1G59990
|
EMB3108, EMBRYO DEFECTIVE 3108, HS3, HEAVY SEED 3, RH22, RNA helicase 22 |
-0.613 |
-0.108 |
| 387 |
264611_at |
AT1G04680
|
[Pectin lyase-like superfamily protein] |
-0.608 |
-0.068 |
| 388 |
254221_at |
AT4G23820
|
[Pectin lyase-like superfamily protein] |
-0.608 |
-0.038 |
| 389 |
261198_at |
AT1G12940
|
ATNRT2.5, nitrate transporter2.5, NRT2.5, nitrate transporter2.5 |
-0.606 |
0.310 |
| 390 |
252971_at |
AT4G38770
|
ATPRP4, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 4, PRP4, proline-rich protein 4 |
-0.605 |
-0.030 |
| 391 |
258920_at |
AT3G10520
|
AHB2, haemoglobin 2, ARATH GLB2, ATGLB2, ARABIDOPSIS HEMOGLOBIN 2, GLB2, HEMOGLOBIN 2, HB2, haemoglobin 2, NSHB2, NON-SYMBIOTIC HAEMOGLOBIN 2 |
-0.603 |
-0.029 |
| 392 |
266300_at |
AT2G01420
|
ATPIN4, ARABIDOPSIS PIN-FORMED 4, PIN4, PIN-FORMED 4 |
-0.601 |
-0.014 |
| 393 |
258156_at |
AT3G18050
|
unknown |
-0.599 |
-0.025 |
| 394 |
258589_at |
AT3G04290
|
ATLTL1, LTL1, Li-tolerant lipase 1 |
-0.598 |
-0.120 |
| 395 |
254110_at |
AT4G25260
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.596 |
0.002 |
| 396 |
266957_at |
AT2G34640
|
HMR, HEMERA, PTAC12, plastid transcriptionally active 12, TAC12 |
-0.595 |
-0.188 |
| 397 |
245555_at |
AT4G15390
|
[HXXXD-type acyl-transferase family protein] |
-0.591 |
-0.014 |
| 398 |
248912_at |
AT5G45670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.590 |
-0.161 |
| 399 |
247383_at |
AT5G63410
|
[Leucine-rich repeat protein kinase family protein] |
-0.590 |
-0.074 |
| 400 |
266996_at |
AT2G34490
|
CYP710A2, cytochrome P450, family 710, subfamily A, polypeptide 2 |
-0.589 |
-0.087 |
| 401 |
253650_at |
AT4G30020
|
[PA-domain containing subtilase family protein] |
-0.588 |
-0.090 |
| 402 |
253270_at |
AT4G34160
|
CYCD3, CYCD3;1, CYCLIN D3;1 |
-0.586 |
-0.115 |
| 403 |
258145_at |
AT3G18200
|
UMAMIT4, Usually multiple acids move in and out Transporters 4 |
-0.585 |
-0.049 |
| 404 |
252537_at |
AT3G45710
|
[Major facilitator superfamily protein] |
-0.584 |
-0.000 |
| 405 |
265586_at |
AT2G19990
|
PR-1-LIKE, pathogenesis-related protein-1-like |
-0.581 |
0.040 |
| 406 |
266395_at |
AT2G43100
|
ATLEUD1, IPMI2, isopropylmalate isomerase 2 |
-0.580 |
-0.257 |
| 407 |
267562_at |
AT2G39670
|
[Radical SAM superfamily protein] |
-0.576 |
-0.073 |
| 408 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
-0.576 |
-0.057 |
| 409 |
260181_at |
AT1G70710
|
ATGH9B1, glycosyl hydrolase 9B1, CEL1, CELLULASE 1, GH9B1, glycosyl hydrolase 9B1 |
-0.576 |
-0.030 |
| 410 |
248427_at |
AT5G51750
|
ATSBT1.3, subtilase 1.3, SBT1.3, subtilase 1.3 |
-0.576 |
-0.087 |
| 411 |
254737_at |
AT4G13840
|
CER26, ECERIFERUM 26 |
-0.573 |
-0.043 |
| 412 |
263836_at |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
-0.569 |
0.362 |
| 413 |
259375_at |
AT3G16370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.569 |
-0.029 |
| 414 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
-0.569 |
-0.174 |
| 415 |
251287_at |
AT3G61820
|
[Eukaryotic aspartyl protease family protein] |
-0.567 |
-0.084 |
| 416 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
-0.566 |
-0.168 |
| 417 |
256655_at |
AT3G18890
|
AtTic62, translocon at the inner envelope membrane of chloroplasts 62, Tic62, translocon at the inner envelope membrane of chloroplasts 62 |
-0.566 |
-0.088 |
| 418 |
251360_at |
AT3G61210
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.563 |
-0.014 |
| 419 |
265704_at |
AT2G03420
|
unknown |
-0.560 |
-0.109 |
| 420 |
256062_at |
AT1G07090
|
LSH6, LIGHT SENSITIVE HYPOCOTYLS 6 |
-0.558 |
-0.022 |
| 421 |
250992_at |
AT5G02260
|
ATEXP9, ATEXPA9, expansin A9, ATHEXP ALPHA 1.10, EXP9, EXPA9, expansin A9 |
-0.557 |
0.001 |
| 422 |
264201_at |
AT1G22630
|
unknown |
-0.557 |
-0.115 |
| 423 |
246108_at |
AT5G28630
|
[glycine-rich protein] |
-0.556 |
0.036 |
| 424 |
253480_at |
AT4G31840
|
AtENODL15, ENODL15, early nodulin-like protein 15 |
-0.556 |
-0.109 |
| 425 |
253871_at |
AT4G27440
|
PORB, protochlorophyllide oxidoreductase B |
-0.555 |
0.004 |
| 426 |
247774_at |
AT5G58660
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.555 |
-0.041 |
| 427 |
249894_at |
AT5G22580
|
[Stress responsive A/B Barrel Domain] |
-0.554 |
-0.059 |
| 428 |
245196_at |
AT1G67750
|
[Pectate lyase family protein] |
-0.552 |
-0.167 |
| 429 |
266801_at |
AT2G22870
|
EMB2001, embryo defective 2001 |
-0.551 |
-0.087 |
| 430 |
266950_at |
AT2G18910
|
[hydroxyproline-rich glycoprotein family protein] |
-0.550 |
0.005 |
| 431 |
259763_at |
AT1G77630
|
LYM3, lysin-motif (LysM) domain protein 3, LYP3, LysM-containing receptor protein 3 |
-0.550 |
-0.159 |
| 432 |
262658_at |
AT1G14220
|
[Ribonuclease T2 family protein] |
-0.549 |
-0.046 |
| 433 |
252343_at |
AT3G48610
|
NPC6, non-specific phospholipase C6 |
-0.549 |
-0.101 |
| 434 |
259373_at |
AT1G69160
|
unknown |
-0.548 |
0.131 |
| 435 |
267034_at |
AT2G38310
|
PYL4, PYR1-like 4, RCAR10, regulatory components of ABA receptor 10 |
-0.548 |
0.042 |
| 436 |
255447_at |
AT4G02790
|
EMB3129, EMBRYO DEFECTIVE 3129 |
-0.547 |
-0.067 |
| 437 |
260982_at |
AT1G53520
|
AtFAP3, FAP3, fatty-acid-binding protein 3 |
-0.546 |
-0.003 |
| 438 |
247337_at |
AT5G63660
|
LCR74, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 74, PDF2.5 |
-0.545 |
-0.020 |
| 439 |
263914_at |
AT2G36400
|
AtGRF3, growth-regulating factor 3, GRF3, growth-regulating factor 3 |
-0.544 |
0.109 |
| 440 |
246562_at |
AT5G15580
|
LNG1, LONGIFOLIA1, TRM2, TON1 Recruiting Motif 2 |
-0.544 |
-0.186 |
| 441 |
257756_at |
AT3G18680
|
[Amino acid kinase family protein] |
-0.543 |
-0.053 |
| 442 |
246505_at |
AT5G16250
|
unknown |
-0.541 |
-0.089 |
| 443 |
259660_at |
AT1G55260
|
LTPG6, glycosylphosphatidylinositol-anchored lipid protein transfer 6 |
-0.539 |
-0.099 |
| 444 |
262324_at |
AT1G64170
|
ATCHX16, cation/H+ exchanger 16, CHX16, cation/H+ exchanger 16 |
-0.538 |
0.344 |
| 445 |
262838_at |
AT1G14960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.537 |
-0.027 |
| 446 |
255277_at |
AT4G04890
|
PDF2, protodermal factor 2 |
-0.537 |
-0.029 |
| 447 |
254233_at |
AT4G23800
|
3xHMG-box2, 3xHigh Mobility Group-box2 |
-0.537 |
-0.242 |
| 448 |
262109_at |
AT1G02730
|
ATCSLD5, cellulose synthase-like D5, CSLD5, CELLULOSE SYNTHASE LIKE D5, CSLD5, cellulose synthase-like D5, SOS6, SALT OVERLY SENSITIVE 6 |
-0.536 |
-0.105 |
| 449 |
262260_at |
AT1G70850
|
MLP34, MLP-like protein 34 |
-0.536 |
-0.050 |
| 450 |
265049_at |
AT1G52060
|
[Mannose-binding lectin superfamily protein] |
-0.535 |
-0.008 |
| 451 |
252215_at |
AT3G50240
|
KICP-02 |
-0.534 |
-0.151 |
| 452 |
259000_at |
AT3G01860
|
unknown |
-0.534 |
-0.050 |
| 453 |
248037_at |
AT5G55930
|
ATOPT1, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 1, OPT1, oligopeptide transporter 1 |
-0.532 |
0.207 |
| 454 |
247216_at |
AT5G64860
|
DPE1, disproportionating enzyme |
-0.532 |
-0.042 |
| 455 |
265588_at |
AT2G19970
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.531 |
-0.048 |
| 456 |
247816_at |
AT5G58260
|
NdhN, NADH dehydrogenase-like complex N |
-0.528 |
-0.113 |
| 457 |
262951_at |
AT1G75500
|
UMAMIT5, Usually multiple acids move in and out Transporters 5, WAT1, Walls Are Thin 1 |
-0.528 |
-0.050 |
| 458 |
259658_at |
AT1G55370
|
NDF5, NDH-dependent cyclic electron flow 5 |
-0.526 |
-0.090 |
| 459 |
247943_at |
AT5G57170
|
[Tautomerase/MIF superfamily protein] |
-0.526 |
-0.038 |
| 460 |
254789_at |
AT4G12880
|
AtENODL19, ENODL19, early nodulin-like protein 19 |
-0.524 |
-0.033 |
| 461 |
253218_at |
AT4G34980
|
SLP2, subtilisin-like serine protease 2 |
-0.523 |
-0.003 |
| 462 |
253233_at |
AT4G34290
|
[SWIB/MDM2 domain superfamily protein] |
-0.521 |
-0.089 |
| 463 |
248854_at |
AT5G46580
|
[pentatricopeptide (PPR) repeat-containing protein] |
-0.516 |
-0.130 |
| 464 |
264900_at |
AT1G23080
|
ATPIN7, ARABIDOPSIS PIN-FORMED 7, PIN7, PIN-FORMED 7 |
-0.514 |
0.017 |
| 465 |
249876_at |
AT5G23060
|
CaS, calcium sensing receptor |
-0.511 |
-0.086 |
| 466 |
251310_at |
AT3G61150
|
HD-GL2-1, HOMEODOMAIN-GLABRA2 1, HDG1, homeodomain GLABROUS 1 |
-0.510 |
0.026 |
| 467 |
266889_at |
AT2G44640
|
unknown |
-0.509 |
-0.035 |
| 468 |
255403_at |
AT4G03400
|
DFL2, DWARF IN LIGHT 2, GH3-10 |
-0.508 |
-0.031 |
| 469 |
262883_at |
AT1G64780
|
AMT1;2, ammonium transporter 1;2, ATAMT1;2, ammonium transporter 1;2 |
-0.507 |
-0.037 |
| 470 |
258480_at |
AT3G02640
|
unknown |
-0.506 |
-0.157 |
| 471 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
-0.506 |
0.032 |
| 472 |
247450_at |
AT5G62350
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.505 |
-0.011 |
| 473 |
260297_at |
AT1G80280
|
[alpha/beta-Hydrolases superfamily protein] |
-0.505 |
-0.110 |
| 474 |
250444_at |
AT5G10560
|
[Glycosyl hydrolase family protein] |
-0.505 |
-0.049 |
| 475 |
249347_at |
AT5G40830
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.505 |
-0.115 |
| 476 |
256774_at |
AT3G13760
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.504 |
-0.009 |
| 477 |
260107_at |
AT1G66430
|
[pfkB-like carbohydrate kinase family protein] |
-0.504 |
-0.019 |
| 478 |
248186_at |
AT5G53880
|
unknown |
-0.504 |
-0.202 |
| 479 |
247914_at |
AT5G57540
|
AtXTH13, XTH13, xyloglucan endotransglucosylase/hydrolase 13 |
-0.503 |
-0.027 |
| 480 |
256237_at |
AT3G12610
|
DRT100, DNA-DAMAGE REPAIR/TOLERATION 100 |
-0.503 |
-0.060 |
| 481 |
265076_at |
AT1G55490
|
CPN60B, chaperonin 60 beta, Cpn60beta1, chaperonin-60beta1, LEN1, LESION INITIATION 1 |
-0.502 |
-0.154 |
| 482 |
246011_at |
AT5G08330
|
AtTCP11, TCP11, TCP domain protein 11 |
-0.501 |
-0.013 |
| 483 |
246911_at |
AT5G25810
|
tny, TINY |
-0.499 |
-0.058 |
| 484 |
246855_at |
AT5G26280
|
[TRAF-like family protein] |
-0.498 |
0.006 |
| 485 |
247474_at |
AT5G62280
|
unknown |
-0.497 |
0.436 |
| 486 |
252048_at |
AT3G52500
|
[Eukaryotic aspartyl protease family protein] |
-0.495 |
-0.059 |
| 487 |
256097_at |
AT1G13670
|
unknown |
-0.495 |
0.066 |
| 488 |
247826_at |
AT5G58480
|
[O-Glycosyl hydrolases family 17 protein] |
-0.494 |
-0.086 |
| 489 |
261055_at |
AT1G01300
|
[Eukaryotic aspartyl protease family protein] |
-0.493 |
-0.016 |
| 490 |
254025_at |
AT4G25790
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.490 |
-0.012 |
| 491 |
252433_at |
AT3G47560
|
[alpha/beta-Hydrolases superfamily protein] |
-0.490 |
-0.127 |
| 492 |
255053_at |
AT4G09730
|
RH39, RH39 |
-0.490 |
-0.143 |
| 493 |
261070_at |
AT1G07390
|
AtRLP1, receptor like protein 1, RLP1, receptor like protein 1 |
-0.490 |
0.025 |
| 494 |
257294_at |
AT3G15570
|
[Phototropic-responsive NPH3 family protein] |
-0.488 |
-0.063 |
| 495 |
249826_at |
AT5G23310
|
FSD3, Fe superoxide dismutase 3 |
-0.487 |
-0.111 |
| 496 |
264689_at |
AT1G09900
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
-0.487 |
-0.112 |
| 497 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
-0.487 |
1.039 |
| 498 |
266215_at |
AT2G06850
|
EXGT-A1, endoxyloglucan transferase A1, EXT, ENDOXYLOGLUCAN TRANSFERASE, XTH4, xyloglucan endotransglucosylase/hydrolase 4 |
-0.486 |
-0.020 |
| 499 |
248201_at |
AT5G54180
|
PTAC15, plastid transcriptionally active 15 |
-0.485 |
-0.117 |
| 500 |
257203_at |
AT3G23730
|
XTH16, xyloglucan endotransglucosylase/hydrolase 16 |
-0.484 |
-0.130 |
| 501 |
254280_at |
AT4G22756
|
ATSMO1-2, SMO1-2, sterol C4-methyl oxidase 1-2 |
-0.483 |
-0.020 |
| 502 |
254251_at |
AT4G23300
|
CRK22, cysteine-rich RLK (RECEPTOR-like protein kinase) 22 |
-0.483 |
0.092 |
| 503 |
256525_at |
AT1G66180
|
[Eukaryotic aspartyl protease family protein] |
-0.482 |
0.095 |
| 504 |
247218_at |
AT5G65010
|
ASN2, asparagine synthetase 2 |
-0.481 |
-0.059 |
| 505 |
253763_at |
AT4G28850
|
ATXTH26, XTH26, xyloglucan endotransglucosylase/hydrolase 26 |
-0.478 |
-0.040 |
| 506 |
266587_at |
AT2G14880
|
[SWIB/MDM2 domain superfamily protein] |
-0.477 |
-0.085 |
| 507 |
251420_at |
AT3G60490
|
[Integrase-type DNA-binding superfamily protein] |
-0.477 |
-0.014 |
| 508 |
257856_at |
AT3G12930
|
[Lojap-related protein] |
-0.475 |
-0.052 |
| 509 |
254783_at |
AT4G12830
|
[alpha/beta-Hydrolases superfamily protein] |
-0.475 |
-0.169 |
| 510 |
247696_at |
AT5G59780
|
ATMYB59, MYB DOMAIN PROTEIN 59, ATMYB59-1, ATMYB59-2, ATMYB59-3, MYB59, myb domain protein 59 |
-0.475 |
-0.196 |
| 511 |
262040_at |
AT1G80080
|
AtRLP17, Receptor Like Protein 17, TMM, TOO MANY MOUTHS |
-0.474 |
0.032 |
| 512 |
262937_at |
AT1G79560
|
EMB1047, EMBRYO DEFECTIVE 1047, EMB156, EMBRYO DEFECTIVE 156, EMB36, EMBRYO DEFECTIVE 36, FTSH12, FTSH protease 12 |
-0.474 |
-0.115 |
| 513 |
249425_at |
AT5G39790
|
5'-AMP-activated protein kinase-related |
-0.473 |
-0.193 |
| 514 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
-0.472 |
-0.135 |
| 515 |
247563_at |
AT5G61130
|
AtPDCB1, PDCB1, plasmodesmata callose-binding protein 1 |
-0.472 |
-0.090 |
| 516 |
255818_at |
AT2G33570
|
GALS1, galactan synthase 1 |
-0.471 |
-0.068 |
| 517 |
262376_at |
AT1G72970
|
EDA17, embryo sac development arrest 17, HTH, HOTHEAD |
-0.470 |
-0.099 |
| 518 |
248961_at |
AT5G45650
|
[subtilase family protein] |
-0.469 |
-0.049 |
| 519 |
257137_at |
AT3G28860
|
ABCB19, ATP-binding cassette B19, ATABCB19, Arabidopsis thaliana ATP-binding cassette B19, ATMDR1, ATMDR11, MULTIDRUG RESISTANCE PROTEIN 11, ATPGP19, MDR1, MDR11, PGP19, P-GLYCOPROTEIN 19 |
-0.468 |
-0.020 |
| 520 |
264319_at |
AT1G04110
|
AtSDD1, SDD1, STOMATAL DENSITY AND DISTRIBUTION 1 |
-0.468 |
0.057 |
| 521 |
256746_at |
AT3G29320
|
PHS1, alpha-glucan phosphorylase 1 |
-0.468 |
-0.110 |
| 522 |
266291_at |
AT2G29320
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.468 |
-0.082 |
| 523 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
-0.467 |
-0.019 |
| 524 |
250952_at |
AT5G03570
|
ATIREG2, ARABIDOPSIS THALIANA IRON-REGULATED PROTEIN 2, FPN2, FERROPORTIN 2, IREG2, iron regulated 2 |
-0.467 |
-0.063 |
| 525 |
255465_at |
AT4G02990
|
BSM, BELAYA SMERT, RUG2, RUGOSA 2 |
-0.467 |
-0.133 |
| 526 |
249162_at |
AT5G42765
|
unknown |
-0.466 |
-0.083 |
| 527 |
253949_at |
AT4G26780
|
AR192, MGE2, mitochondrial GrpE 2 |
-0.464 |
-0.020 |
| 528 |
265884_at |
AT2G42320
|
[nucleolar protein gar2-related] |
-0.462 |
-0.129 |
| 529 |
255623_at |
AT4G01310
|
[Ribosomal L5P family protein] |
-0.462 |
-0.029 |
| 530 |
262086_at |
AT1G56050
|
[GTP-binding protein-related] |
-0.460 |
-0.163 |
| 531 |
248095_at |
AT5G55230
|
ATMAP65-1, microtubule-associated proteins 65-1, MAP65-1, MAP65-1, microtubule-associated proteins 65-1 |
-0.459 |
-0.098 |
| 532 |
255632_at |
AT4G00680
|
ADF8, actin depolymerizing factor 8 |
-0.458 |
-0.032 |
| 533 |
264790_at |
AT2G17820
|
AHK1, ATHK1, histidine kinase 1, HK1, histidine kinase 1 |
-0.457 |
-0.057 |
| 534 |
264069_at |
AT2G28000
|
CH-CPN60A, CHLOROPLAST CHAPERONIN 60ALPHA, CPN60A, chaperonin-60alpha, Cpn60alpha1, chaperonin-60alpha1, SLP, SCHLEPPERLESS |
-0.456 |
-0.083 |
| 535 |
265336_at |
AT2G18290
|
APC10, anaphase promoting complex 10, AtAPC10, EMB2783, EMBRYO DEFECTIVE 2783 |
-0.456 |
-0.045 |
| 536 |
249026_at |
AT5G44785
|
OSB3, organellar single-stranded DNA binding protein 3 |
-0.455 |
-0.104 |
| 537 |
258764_at |
AT3G10720
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.454 |
0.034 |
| 538 |
263386_at |
AT2G40150
|
TBL28, TRICHOME BIREFRINGENCE-LIKE 28 |
-0.454 |
-0.106 |
| 539 |
260451_at |
AT1G72360
|
AtERF73, ERF73, ethylene response factor 73, HRE1, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 1 |
-0.454 |
-0.014 |
| 540 |
263970_at |
AT2G42850
|
CYP718, cytochrome P450, family 718 |
-0.454 |
0.055 |
| 541 |
255104_at |
AT4G08685
|
SAH7 |
-0.453 |
-0.047 |
| 542 |
262543_at |
AT1G34245
|
EPF2, EPIDERMAL PATTERNING FACTOR 2 |
-0.453 |
0.164 |
| 543 |
247037_at |
AT5G67070
|
RALFL34, ralf-like 34 |
-0.449 |
-0.081 |
| 544 |
262230_at |
AT1G68560
|
ATXYL1, ALPHA-XYLOSIDASE 1, AXY3, altered xyloglucan 3, TRG1, thermoinhibition resistant germination 1, XYL1, alpha-xylosidase 1 |
-0.448 |
-0.006 |
| 545 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
-0.447 |
-0.072 |
| 546 |
248420_at |
AT5G51560
|
[Leucine-rich repeat protein kinase family protein] |
-0.447 |
-0.129 |
| 547 |
249354_at |
AT5G40480
|
EMB3012, embryo defective 3012 |
-0.447 |
-0.157 |
| 548 |
258355_at |
AT3G14330
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.446 |
-0.164 |
| 549 |
259680_at |
AT1G77690
|
LAX3, like AUX1 3 |
-0.445 |
0.001 |
| 550 |
259161_at |
AT3G01500
|
ATBCA1, BETA CARBONIC ANHYDRASE 1, ATSABP3, ARABIDOPSIS THALIANA SALICYLIC ACID-BINDING PROTEIN 3, CA1, carbonic anhydrase 1, SABP3, SALICYLIC ACID-BINDING PROTEIN 3 |
-0.444 |
-0.022 |
| 551 |
256676_at |
AT3G52180
|
ATPTPKIS1, ATSEX4, DSP4, DUAL-SPECIFICITY PROTEIN PHOSPHATASE 4, SEX4, STARCH-EXCESS 4 |
-0.443 |
-0.093 |
| 552 |
253278_at |
AT4G34220
|
[Leucine-rich repeat protein kinase family protein] |
-0.442 |
-0.031 |
| 553 |
247116_at |
AT5G65970
|
ATMLO10, MILDEW RESISTANCE LOCUS O 10, MLO10, MILDEW RESISTANCE LOCUS O 10 |
-0.441 |
-0.067 |
| 554 |
266668_at |
AT2G29760
|
OTP81, ORGANELLE TRANSCRIPT PROCESSING 81 |
-0.440 |
-0.060 |
| 555 |
245845_at |
AT1G26150
|
AtPERK10, proline-rich extensin-like receptor kinase 10, PERK10, proline-rich extensin-like receptor kinase 10 |
-0.438 |
-0.069 |
| 556 |
255516_at |
AT4G02270
|
RHS13, root hair specific 13 |
-0.437 |
0.043 |
| 557 |
264497_at |
AT1G30840
|
ATPUP4, purine permease 4, PUP4, purine permease 4 |
-0.437 |
-0.016 |
| 558 |
258552_at |
AT3G07010
|
[Pectin lyase-like superfamily protein] |
-0.437 |
-0.051 |
| 559 |
251956_at |
AT3G53460
|
CP29, chloroplast RNA-binding protein 29 |
-0.436 |
-0.021 |
| 560 |
253244_at |
AT4G34580
|
AtSFH1, COW1, CAN OF WORMS1, SRH1, SHORT ROOT HAIR 1 |
-0.436 |
0.074 |
| 561 |
264343_at |
AT1G11850
|
unknown |
-0.435 |
-0.012 |
| 562 |
256452_at |
AT1G75240
|
AtHB33, homeobox protein 33, HB33, homeobox protein 33, ZHD5, zinc-finger homeodomain 5 |
-0.434 |
0.020 |
| 563 |
251374_at |
AT3G60390
|
HAT3, homeobox-leucine zipper protein 3 |
-0.433 |
0.081 |
| 564 |
263053_at |
AT2G13440
|
[glucose-inhibited division family A protein] |
-0.433 |
-0.089 |
| 565 |
261762_at |
AT1G15510
|
ATECB2, ARABIDOPSIS EARLY CHLOROPLAST BIOGENESIS2, ECB2, EARLY CHLOROPLAST BIOGENESIS2, VAC1, VANILLA CREAM 1 |
-0.432 |
-0.189 |
| 566 |
256168_at |
AT1G51805
|
[Leucine-rich repeat protein kinase family protein] |
-0.432 |
-0.059 |
| 567 |
249718_at |
AT5G35740
|
[Carbohydrate-binding X8 domain superfamily protein] |
-0.431 |
-0.093 |
| 568 |
260876_at |
AT1G21460
|
AtSWEET1, SWEET1 |
-0.431 |
-0.102 |
| 569 |
245150_at |
AT2G47590
|
PHR2, photolyase/blue-light receptor 2 |
-0.431 |
-0.078 |
| 570 |
261611_at |
AT1G49730
|
[Protein kinase superfamily protein] |
-0.430 |
-0.054 |
| 571 |
258859_at |
AT3G02120
|
[hydroxyproline-rich glycoprotein family protein] |
-0.430 |
-0.170 |
| 572 |
262728_at |
AT1G75820
|
ATCLV1, CLV1, CLAVATA 1, FAS3, FASCIATA 3, FLO5, FLOWER DEVELOPMENT 5 |
-0.429 |
-0.036 |
| 573 |
267591_at |
AT2G39705
|
DVL11, DEVIL 11, RTFL8, ROTUNDIFOLIA like 8 |
-0.429 |
0.040 |
| 574 |
253483_at |
AT4G31910
|
BAT1, BR-related AcylTransferase1, PIZ, PIZZA |
-0.428 |
0.001 |
| 575 |
254133_at |
AT4G24810
|
[Protein kinase superfamily protein] |
-0.425 |
-0.154 |
| 576 |
245859_at |
AT5G28290
|
ATNEK3, NIMA-related kinase 3, NEK3, NIMA-related kinase 3 |
-0.425 |
-0.041 |
| 577 |
250529_at |
AT5G08610
|
PDE340, PIGMENT DEFECTIVE 340 |
-0.424 |
-0.053 |
| 578 |
258895_at |
AT3G05600
|
[alpha/beta-Hydrolases superfamily protein] |
-0.424 |
0.066 |
| 579 |
256383_at |
AT1G66820
|
[glycine-rich protein] |
-0.423 |
-0.098 |
| 580 |
265066_at |
AT1G03870
|
FLA9, FASCICLIN-like arabinoogalactan 9 |
-0.423 |
-0.100 |
| 581 |
258149_at |
AT3G18110
|
EMB1270, embryo defective 1270 |
-0.422 |
-0.134 |
| 582 |
261301_at |
AT1G48570
|
[zinc finger (Ran-binding) family protein] |
-0.421 |
-0.032 |
| 583 |
263674_at |
AT2G04790
|
unknown |
-0.421 |
-0.168 |
| 584 |
248151_at |
AT5G54270
|
LHCB3, light-harvesting chlorophyll B-binding protein 3, LHCB3*1 |
-0.421 |
0.007 |
| 585 |
253749_at |
AT4G29080
|
IAA27, indole-3-acetic acid inducible 27, PAP2, phytochrome-associated protein 2 |
-0.418 |
-0.062 |
| 586 |
248807_at |
AT5G47500
|
PME5, pectin methylesterase 5 |
-0.418 |
-0.145 |
| 587 |
265961_at |
AT2G37400
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.417 |
-0.045 |
| 588 |
259091_at |
AT3G04890
|
[Uncharacterized conserved protein (DUF2358)] |
-0.416 |
-0.016 |
| 589 |
259172_at |
AT3G03630
|
CS26, cysteine synthase 26 |
-0.415 |
-0.052 |
| 590 |
248380_at |
AT5G51820
|
ATPGMP, ARABIDOPSIS THALIANA PHOSPHOGLUCOMUTASE, PGM, phosphoglucomutase, PGM1, STF1, STARCH-FREE 1 |
-0.414 |
-0.094 |
| 591 |
257236_at |
AT3G15095
|
HCF243, high chlorophyll fluorescence 243 |
-0.413 |
-0.064 |
| 592 |
254669_at |
AT4G18370
|
DEG5, degradation of periplasmic proteins 5, DEGP5, DEGP PROTEASE 5, HHOA, PROTEASE HHOA PRECUSOR |
-0.413 |
-0.089 |
| 593 |
245877_at |
AT1G26220
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.411 |
-0.092 |
| 594 |
256675_at |
AT3G52170
|
[DNA binding] |
-0.411 |
-0.202 |
| 595 |
254466_at |
AT4G20430
|
[Subtilase family protein] |
-0.411 |
-0.078 |
| 596 |
262657_at |
AT1G14210
|
[Ribonuclease T2 family protein] |
-0.410 |
0.001 |
| 597 |
247541_at |
AT5G61660
|
[glycine-rich protein] |
-0.409 |
-0.021 |
| 598 |
251497_at |
AT3G59060
|
PIF5, PHYTOCHROME-INTERACTING FACTOR 5, PIL6, phytochrome interacting factor 3-like 6 |
-0.409 |
0.025 |
| 599 |
256784_at |
AT3G13674
|
unknown |
-0.409 |
-0.016 |
| 600 |
261230_at |
AT1G20010
|
TUB5, tubulin beta-5 chain |
-0.407 |
-0.031 |