|
probeID |
AGICode |
Annotation |
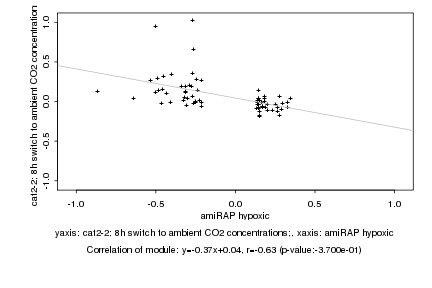
Log2 signal ratio
amiRAP hypoxic |
Log2 signal ratio
cat2-2; 8h switch to ambient CO2 concentrations; |
| 1 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
0.341 |
0.047 |
| 2 |
244973_at |
ATCG00690
|
PSBT, photosystem II reaction center protein T, PSBTC |
0.327 |
-0.070 |
| 3 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
0.326 |
-0.012 |
| 4 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
0.290 |
-0.022 |
| 5 |
244933_at |
ATCG01070
|
NDHE |
0.284 |
-0.095 |
| 6 |
244965_at |
ATCG00590
|
ORF31 |
0.276 |
-0.171 |
| 7 |
264261_at |
AT1G09240
|
ATNAS3, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 3, NAS3, nicotianamine synthase 3 |
0.273 |
0.067 |
| 8 |
255064_at |
AT4G08950
|
EXO, EXORDIUM |
0.260 |
-0.067 |
| 9 |
244970_at |
ATCG00660
|
RPL20, ribosomal protein L20 |
0.260 |
-0.117 |
| 10 |
265722_at |
AT2G40100
|
LHCB4.3, light harvesting complex photosystem II |
0.246 |
-0.029 |
| 11 |
244934_at |
ATCG01080
|
NDHG |
0.230 |
-0.113 |
| 12 |
256569_at |
AT3G19550
|
unknown |
0.199 |
-0.108 |
| 13 |
249726_at |
AT5G35480
|
unknown |
0.198 |
-0.029 |
| 14 |
245018_at |
ATCG00520
|
YCF4 |
0.184 |
-0.070 |
| 15 |
259560_at |
AT1G21270
|
WAK2, wall-associated kinase 2 |
0.180 |
0.049 |
| 16 |
251642_at |
AT3G57520
|
AtSIP2, seed imbibition 2, RS2, raffinose synthase 2, SIP2, seed imbibition 2 |
0.178 |
0.064 |
| 17 |
265404_at |
AT2G16730
|
BGAL13, beta-galactosidase 13 |
0.178 |
0.007 |
| 18 |
255080_at |
AT4G09030
|
AGP10, arabinogalactan protein 10, ATAGP10 |
0.168 |
-0.053 |
| 19 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
0.167 |
-0.072 |
| 20 |
254785_at |
AT4G12730
|
FLA2, FASCICLIN-like arabinogalactan 2 |
0.165 |
-0.057 |
| 21 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
0.163 |
-0.006 |
| 22 |
250980_at |
AT5G03130
|
unknown |
0.149 |
-0.185 |
| 23 |
267076_at |
AT2G41090
|
[Calcium-binding EF-hand family protein] |
0.148 |
0.014 |
| 24 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
0.147 |
-0.121 |
| 25 |
267123_at |
AT2G23560
|
ATMES7, ARABIDOPSIS THALIANA METHYL ESTERASE 7, MES7, methyl esterase 7 |
0.146 |
-0.176 |
| 26 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
0.146 |
0.025 |
| 27 |
262947_at |
AT1G75750
|
GASA1, GAST1 protein homolog 1 |
0.144 |
-0.030 |
| 28 |
255808_at |
AT4G10280
|
[RmlC-like cupins superfamily protein] |
0.143 |
-0.081 |
| 29 |
266395_at |
AT2G43100
|
ATLEUD1, IPMI2, isopropylmalate isomerase 2 |
0.143 |
-0.032 |
| 30 |
253617_at |
AT4G30410
|
[sequence-specific DNA binding transcription factors] |
0.142 |
-0.070 |
| 31 |
257017_at |
AT3G19620
|
[Glycosyl hydrolase family protein] |
0.142 |
0.039 |
| 32 |
258452_at |
AT3G22370
|
AOX1A, alternative oxidase 1A, ATAOX1A, AtHSR3, HSR3, hyper-sensitivity-related 3 |
0.141 |
0.141 |
| 33 |
249314_at |
AT5G41180
|
[leucine-rich repeat transmembrane protein kinase family protein] |
0.141 |
0.040 |
| 34 |
246371_at |
AT1G51940
|
LYK3, LysM-containing receptor-like kinase 3 |
0.136 |
-0.007 |
| 35 |
251906_at |
AT3G53720
|
ATCHX20, cation/H+ exchanger 20, CHX20, cation/H+ exchanger 20 |
0.135 |
0.013 |
| 36 |
262179_at |
AT1G77980
|
AGL66, AGAMOUS-like 66 |
0.133 |
-0.027 |
| 37 |
267532_at |
AT2G42040
|
unknown |
0.129 |
-0.088 |
| 38 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
-0.870 |
0.137 |
| 39 |
253505_at |
AT4G31970
|
CYP82C2, cytochrome P450, family 82, subfamily C, polypeptide 2 |
-0.647 |
0.046 |
| 40 |
254832_at |
AT4G12490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.535 |
0.273 |
| 41 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
-0.505 |
0.949 |
| 42 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
-0.503 |
0.118 |
| 43 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
-0.495 |
0.298 |
| 44 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
-0.484 |
0.149 |
| 45 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
-0.468 |
-0.014 |
| 46 |
267567_at |
AT2G30770
|
CYP71A13, cytochrome P450, family 71, subfamily A, polypeptide 13 |
-0.461 |
0.161 |
| 47 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
-0.453 |
0.324 |
| 48 |
260101_at |
AT1G73260
|
ATKTI1, ARABIDOPSIS THALIANA KUNITZ TRYPSIN INHIBITOR 1, KTI1, kunitz trypsin inhibitor 1 |
-0.437 |
0.107 |
| 49 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
-0.410 |
-0.001 |
| 50 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.406 |
0.353 |
| 51 |
259040_at |
AT3G09270
|
ATGSTU8, glutathione S-transferase TAU 8, GSTU8, glutathione S-transferase TAU 8 |
-0.344 |
0.202 |
| 52 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
-0.331 |
0.019 |
| 53 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
-0.322 |
0.053 |
| 54 |
258941_at |
AT3G09940
|
ATMDAR3, ARABIDOPSIS THALIANA MONODEHYDROASCORBATE REDUCTASE 3, MDAR2, MONODEHYDROASCORBATE REDUCTASE 2, MDAR3, MONODEHYDROASCORBATE REDUCTASE 3, MDHAR, monodehydroascorbate reductase |
-0.318 |
0.117 |
| 55 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
-0.318 |
0.137 |
| 56 |
256933_at |
AT3G22600
|
LTPG5, glycosylphosphatidylinositol-anchored lipid protein transfer 5 |
-0.318 |
0.191 |
| 57 |
247224_at |
AT5G65080
|
AGL68, AGAMOUS-like 68, MAF5, MADS AFFECTING FLOWERING 5 |
-0.313 |
-0.048 |
| 58 |
258957_at |
AT3G01420
|
ALPHA-DOX1, alpha-dioxygenase 1, DIOX1, DOX1, PADOX-1, plant alpha dioxygenase 1 |
-0.305 |
0.044 |
| 59 |
258203_at |
AT3G13950
|
unknown |
-0.291 |
0.207 |
| 60 |
245393_at |
AT4G16260
|
[Glycosyl hydrolase superfamily protein] |
-0.282 |
0.191 |
| 61 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
-0.272 |
0.357 |
| 62 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
-0.271 |
1.035 |
| 63 |
253799_at |
AT4G28140
|
[Integrase-type DNA-binding superfamily protein] |
-0.270 |
0.066 |
| 64 |
267391_at |
AT2G44480
|
BGLU17, beta glucosidase 17 |
-0.265 |
-0.020 |
| 65 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
-0.264 |
0.661 |
| 66 |
261420_at |
AT1G07720
|
KCS3, 3-ketoacyl-CoA synthase 3 |
-0.257 |
0.008 |
| 67 |
248218_at |
AT5G53710
|
unknown |
-0.255 |
-0.008 |
| 68 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
-0.251 |
0.278 |
| 69 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
-0.244 |
0.143 |
| 70 |
265418_at |
AT2G20880
|
AtERF53, ERF53, ERF domain 53 |
-0.231 |
0.024 |
| 71 |
245593_at |
AT4G14550
|
IAA14, indole-3-acetic acid inducible 14, SLR, SOLITARY ROOT |
-0.219 |
-0.004 |
| 72 |
253962_at |
AT4G26460
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.218 |
-0.058 |
| 73 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
-0.218 |
0.274 |